Constraints of Viral RNA Synthesis on Codon Usage of Negative-Strand RNA Virus
- PMID: 30541832
- PMCID: PMC6384081
- DOI: 10.1128/JVI.01775-18
Constraints of Viral RNA Synthesis on Codon Usage of Negative-Strand RNA Virus
Abstract
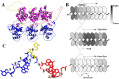
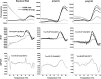
Negative-strand RNA viruses (NSVs) include some of the most pathogenic human viruses known. NSVs completely rely on the host cell for protein translation, but their codon usage bias is often different from that of the host. This discrepancy may have originated from the unique mechanism of NSV RNA synthesis in that the genomic RNA sequestered in the nucleocapsid serves as the template. The stability of the genomic RNA in the nucleocapsid appears to regulate its accessibility to the viral RNA polymerase, thus placing constraints on codon usage to balance viral RNA synthesis. By in situ analyses of vesicular stomatitis virus RNA synthesis, specific activities of viral RNA synthesis were correlated with the genomic RNA sequence. It was found that by simply altering the sequence and not the amino acid that it encoded, a significant reduction, up to an ∼750-fold reduction, in viral RNA transcripts occurred. Through subsequent sequence analysis and thermal shift assays, it was found that the purine/pyrimidine content modulates the overall stability of the polymerase complex, resulting in alteration of the activity of viral RNA synthesis. The codon usage is therefore constrained by the obligation of the NSV genome for viral RNA synthesis.IMPORTANCE Negative-strand RNA viruses (NSVs) include the most pathogenic viruses known. New methods to monitor their evolutionary trends are urgently needed for the development of antivirals and vaccines. The protein translation machinery of the host cell is currently recognized as a main genomic regulator of RNA virus evolution, which works especially well for positive-strand RNA viruses. However, this approach fails for NSVs because it does not consider the unique mechanism of their viral RNA synthesis. For NSVs, the viral RNA-dependent RNA polymerase (vRdRp) must gain access to the genome sequestered in the nucleocapsid. Our work suggests a paradigm shift that the interactions between the RNA genome and the nucleocapsid protein regulate the activity of vRdRp, which selects codon usage.
Keywords: nucleocapsid; sequestered genome; viral polymerase.
Copyright © 2019 American Society for Microbiology.
Figures


Similar articles
-
Nucleocapsid Structure of Negative Strand RNA Virus.Viruses. 2020 Jul 30;12(8):835. doi: 10.3390/v12080835. Viruses. 2020. PMID: 32751700 Free PMC article. Review.
-
Complementary Mutations in the N and L Proteins for Restoration of Viral RNA Synthesis.J Virol. 2018 Oct 29;92(22):e01417-18. doi: 10.1128/JVI.01417-18. Print 2018 Nov 15. J Virol. 2018. PMID: 30135126 Free PMC article.
-
Second-site mutations selected in transcriptional regulatory sequences compensate for engineered mutations in the vesicular stomatitis virus nucleocapsid protein.J Virol. 2012 Oct;86(20):11266-75. doi: 10.1128/JVI.01238-12. Epub 2012 Aug 8. J Virol. 2012. PMID: 22875970 Free PMC article.
-
The nucleocapsid of vesicular stomatitis virus.Sci China Life Sci. 2012 Apr;55(4):291-300. doi: 10.1007/s11427-012-4307-x. Epub 2012 May 9. Sci China Life Sci. 2012. PMID: 22566085 Review.
-
A Polyamide Inhibits Replication of Vesicular Stomatitis Virus by Targeting RNA in the Nucleocapsid.J Virol. 2018 Mar 28;92(8):e00146-18. doi: 10.1128/JVI.00146-18. Print 2018 Apr 15. J Virol. 2018. PMID: 29437970 Free PMC article.
Cited by
-
Prediction of two novel overlapping ORFs in the genome of SARS-CoV-2.Virology. 2021 Oct;562:149-157. doi: 10.1016/j.virol.2021.07.011. Epub 2021 Jul 28. Virology. 2021. PMID: 34339929 Free PMC article.
-
Nucleocapsid Structure of Negative Strand RNA Virus.Viruses. 2020 Jul 30;12(8):835. doi: 10.3390/v12080835. Viruses. 2020. PMID: 32751700 Free PMC article. Review.
-
The Codon Usage Code for Cotranslational Folding of Viral Capsids.Genome Biol Evol. 2021 Sep 1;13(9):evab089. doi: 10.1093/gbe/evab089. Genome Biol Evol. 2021. PMID: 33914886 Free PMC article.
-
Viral N6-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus.Nat Commun. 2019 Oct 9;10(1):4595. doi: 10.1038/s41467-019-12504-y. Nat Commun. 2019. PMID: 31597913 Free PMC article.
-
Immune Pressure on Polymorphous Influenza B Populations Results in Diverse Hemagglutinin Escape Mutants and Lineage Switching.Vaccines (Basel). 2020 Mar 11;8(1):125. doi: 10.3390/vaccines8010125. Vaccines (Basel). 2020. PMID: 32168968 Free PMC article.
References
-
- Bont L, Checchia PA, Fauroux B, Figueras-Aloy J, Manzoni P, Paes B, Simões EAF, Carbonell-Estrany X. 2016. Defining the epidemiology and burden of severe respiratory syncytial virus infection among infants and children in Western countries. Infect Dis Ther 5:271–298. doi:10.1007/s40121-016-0123-0. - DOI - PMC - PubMed
-
- Wu W, Liu S. 2017. The drug targets and antiviral molecules for treatment of Ebola virus infection. Curr Top Med Chem 17:361–370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

