Hantavirus RdRp Requires a Host Cell Factor for Cap Snatching
- PMID: 30541836
- PMCID: PMC6384069
- DOI: 10.1128/JVI.02088-18
Hantavirus RdRp Requires a Host Cell Factor for Cap Snatching
Abstract
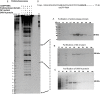
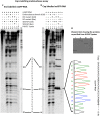
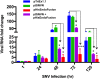
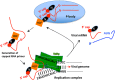
The hantavirus RNA-dependent RNA polymerase (RdRp) snatches 5' capped mRNA fragments from the host cell transcripts and uses them as primers to initiate transcription and replication of the viral genome in the cytoplasm of infected cells. Hantavirus nucleocapsid protein (N protein) binds to the 5' caps of host cell mRNA and protects them from the attack of cellular decapping machinery. N protein rescues long capped mRNA fragments in cellular P bodies that are later processed by an unknown mechanism to generate 10- to 14-nucleotide-long capped RNA primers with a 3' G residue. Hantavirus RdRp has an N-terminal endonuclease domain and a C-terminal uncharacterized domain that harbors a binding site for the N protein. The purified endonuclease domain of RdRp nonspecifically degraded RNA in vitro It is puzzling how such nonspecific endonuclease activity generates primers of appropriate length and specificity during cap snatching. We fused the N-terminal endonuclease domain with the C-terminal uncharacterized domain of the RdRp. The resulting NC mutant, with the assistance of N protein, generated capped primers of appropriate length and specificity from a test mRNA in cells. Bacterially expressed and purified NC mutant and N protein required further incubation with the lysates of human umbilical vein endothelial cells (HUVECs) for the specific endonucleolytic cleavage of a test mRNA to generate capped primers of appropriate length and defined 3' terminus in vitro Our results suggest that an unknown host cell factor facilitates the interaction between N protein and NC mutant and brings the N protein-bound capped RNA fragments in close proximity to the endonuclease domain of the RdRp for specific cleavage at a precise length from the 5' cap. These studies provide critical insights into the cap-snatching mechanism of cytoplasmic viruses and have revealed potential new targets for their therapeutic intervention.IMPORTANCE Humans acquire hantavirus infection by the inhalation of aerosolized excreta of infected rodent hosts. Hantavirus infections cause hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS), with mortality rates of 15% and 50%, respectively (1). Annually 150,000 to 200,000 cases of hantavirus infections are reported worldwide, for which there is no treatment at present. Cap snatching is an early event in the initiation of virus replication in infected hosts. Interruption in cap snatching will inhibit virus replication and will likely improve the prognosis of the hantavirus disease. Our studies provide mechanistic insight into the cap-snatching mechanism and demonstrate the requirement of a host cell factor for successful cap snatching. Identification of this host cell factor will reveal a novel therapeutic target for combating this viral illness.
Keywords: cap snatching; hantavirus; negative-strand RNA virus; nucleocapsid.
Copyright © 2019 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

