New Isolates of Pandoraviruses: Contribution to the Study of Replication Cycle Steps
- PMID: 30541841
- PMCID: PMC6384056
- DOI: 10.1128/JVI.01942-18
New Isolates of Pandoraviruses: Contribution to the Study of Replication Cycle Steps
Abstract
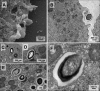
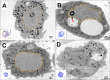
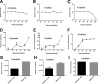
Giant viruses are complex members of the virosphere, exhibiting outstanding structural and genomic features. Among these viruses, the pandoraviruses are some of the most intriguing members, exhibiting giant particles and genomes presenting at up to 2.5 Mb, with many genes having no known function. In this work, we analyzed, by virological and microscopic methods, the replication cycle steps of three new pandoravirus isolates from samples collected in different regions of Brazil. Our data indicate that all analyzed pandoravirus isolates can deeply modify the Acanthamoeba cytoplasmic environment, recruiting mitochondria and membranes into and around the electron-lucent viral factories. We also observed that the viral factories start forming before the complete degradation of the cellular nucleus. Various patterns of pandoravirus particle morphogenesis were observed, and the assembly of the particles seemed to be started either by the apex or by the opposite side. On the basis of the counting of viral particles during the infection time course, we observed that pandoravirus particles could undergo exocytosis after their morphogenesis in a process that involved intense recruitment of membranes that wrapped the just-formed particles. The treatment of infected cells with brefeldin affected particle exocytosis in two of the three analyzed strains, indicating biological variability among isolates. Despite such particle exocytosis, the lysis of host cells also contributed to viral release. This work reinforces knowledge of and reveals important steps in the replication cycle of pandoraviruses.IMPORTANCE The emerging Pandoraviridae family is composed of some of the most complex viruses known to date. Only a few pandoravirus isolates have been described until now, and many aspects of their life cycle remain to be elucidated. A comprehensive description of the replication cycle is pivotal to a better understanding of the biology of the virus. For this report, we describe new pandoraviruses and used different methods to better characterize the steps of the replication cycle of this new group of viruses. Our results provide new information about the diversity and biology of these giant viruses.
Keywords: giant virus; pandoravirus; replication cycle; viral morphogenesis; viral release; virus diversity.
Copyright © 2019 American Society for Microbiology.
Figures






Similar articles
-
In-depth analysis of the replication cycle of Orpheovirus.Virol J. 2019 Dec 16;16(1):158. doi: 10.1186/s12985-019-1268-8. Virol J. 2019. PMID: 31842897 Free PMC article.
-
A Puzzling Anomaly in the 4-Mer Composition of the Giant Pandoravirus Genomes Reveals a Stringent New Evolutionary Selection Process.J Virol. 2019 Nov 13;93(23):e01206-19. doi: 10.1128/JVI.01206-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534042 Free PMC article.
-
Cedratvirus getuliensis replication cycle: an in-depth morphological analysis.Sci Rep. 2018 Mar 5;8(1):4000. doi: 10.1038/s41598-018-22398-3. Sci Rep. 2018. PMID: 29507337 Free PMC article.
-
A strange endocytobiont revealed as largest virus.Curr Opin Microbiol. 2016 Jun;31:58-62. doi: 10.1016/j.mib.2016.02.005. Epub 2016 Mar 24. Curr Opin Microbiol. 2016. PMID: 27016694 Review.
-
Viruses in close associations with free-living amoebae.Parasitol Res. 2015 Nov;114(11):3959-67. doi: 10.1007/s00436-015-4731-5. Epub 2015 Sep 16. Parasitol Res. 2015. PMID: 26374538 Review.
Cited by
-
A history of over 40 years of potentially pathogenic free-living amoeba studies in Brazil - a systematic review.Mem Inst Oswaldo Cruz. 2022 Jul 1;117:e210373. doi: 10.1590/0074-02760210373. eCollection 2022. Mem Inst Oswaldo Cruz. 2022. PMID: 35792751 Free PMC article.
-
In-depth analysis of the replication cycle of Orpheovirus.Virol J. 2019 Dec 16;16(1):158. doi: 10.1186/s12985-019-1268-8. Virol J. 2019. PMID: 31842897 Free PMC article.
-
A long-term prospecting study on giant viruses in terrestrial and marine Brazilian biomes.Virol J. 2024 Jun 10;21(1):135. doi: 10.1186/s12985-024-02404-z. Virol J. 2024. PMID: 38858684 Free PMC article.
-
Yaravirus: A novel 80-nm virus infecting Acanthamoeba castellanii.Proc Natl Acad Sci U S A. 2020 Jul 14;117(28):16579-16586. doi: 10.1073/pnas.2001637117. Epub 2020 Jun 29. Proc Natl Acad Sci U S A. 2020. PMID: 32601223 Free PMC article.
-
A Brief History of Giant Viruses' Studies in Brazilian Biomes.Viruses. 2022 Jan 19;14(2):191. doi: 10.3390/v14020191. Viruses. 2022. PMID: 35215784 Free PMC article. Review.
References
-
- Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106:21848–21853. doi:10.1073/pnas.0911354106. - DOI - PMC - PubMed
-
- Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Coute Y, Rivkina E, Abergel C, Claverie JM. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci U S A 111:4274–4279. doi:10.1073/pnas.1320670111. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

