Iterons Homologous to Helper Geminiviruses Are Essential for Efficient Replication of Betasatellites
- PMID: 30541843
- PMCID: PMC6384059
- DOI: 10.1128/JVI.01532-18
Iterons Homologous to Helper Geminiviruses Are Essential for Efficient Replication of Betasatellites
Abstract
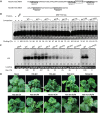
Betasatellites associated with geminiviruses can be replicated promiscuously by distinct geminiviruses but exhibit a preference for cognate helper viruses. However, the cis elements responsible for betasatellite origin recognition have not been characterized. In this study, we identified an iteron-like repeated sequence motif, 5'-GAGGACC-3', in a tobacco curly shoot betasatellite (TbCSB) associated with tobacco curly shoot virus (TbCSV). Competitive DNA binding assays revealed that two core repeats (5'-GGACC-3') are required for specific binding to TbCSV Rep; TbCSB iteron mutants accumulated to greatly reduced levels and lost the cognate helper-mediated replication preference. Interestingly, TbCSV also contains identical repeated sequences that are essential for specific Rep binding and in vivo replication. In order to gain insight into the mechanism by which TbCSB has acquired the cognate iterons, we performed a SELEX (systematic evolution of ligands by exponential enrichment) assay to identify the high-affinity Rep binding ligands from a large pool of randomized sequences. Analysis of SELEX winners showed that all of the sequences contained at least one core iteron-like motif, suggesting that TbCSB has evolved to contain cognate iterons for high-affinity Rep binding. Further analyses of various betasatellite sequences revealed a region upstream of the satellite conserved region replete with iterative sequence motifs, including species-specific repeats and a general repeat (5'-GGTAAAT-3'). Remarkably, the species-specific repeats in many betasatellites are homologous to those in their respective cognate helper begomoviruses, whereas the general repeat is widespread in most of the betasatellite molecules analyzed. These data, taken together, suggest that many betasatellites have evolved to acquire homologous iteron-like sequences for efficient replication mediated by cognate helper viruses.IMPORTANCE The geminivirus-encoded replication initiator protein (Rep) binds to repeated sequence elements (also known as iterons) in the origin of replication that serve as essential cis elements for specific viral replication. Betasatellites associated with begomoviruses can be replicated by cognate or noncognate helper viruses, but the cis elements responsible for betasatellite origin recognition have not been characterized. Using a betasatellite (TbCSB) associated with tobacco curly shoot virus (TbCSV) as a model, we identify two tandem repeats (iterons) in the Rep-binding motif (RBM) that are required for specific Rep binding and efficient replication, and we show that identical iteron sequences present in TbCSV are also necessary for Rep binding and the replication of helper viruses. Extensive analysis of begomovirus/betasatellite sequences shows that many betasatellites contain iteron-like elements homologous to those of their respective cognate helper begomoviruses. Our data suggest that many betasatellites have evolved to acquire homologous iteron-like sequences for efficient replication mediated by cognate helper viruses.
Keywords: betasatellite; geminivirus; iterons; replication; tobacco curly shoot virus.
Copyright © 2019 American Society for Microbiology.
Figures





References
-
- Jose J, Usha R. 2003. Bhendi yellow vein mosaic disease in India is caused by association of a DNA β satellite with a begomovirus. Virology 305:310–317. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

