The Potato Virus X TGBp2 Protein Plays Dual Functional Roles in Viral Replication and Movement
- PMID: 30541845
- PMCID: PMC6384063
- DOI: 10.1128/JVI.01635-18
The Potato Virus X TGBp2 Protein Plays Dual Functional Roles in Viral Replication and Movement
Abstract
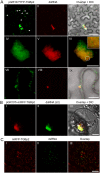
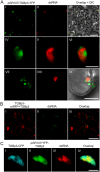
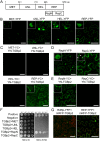
Plant viruses usually encode one or more movement proteins (MP) to accomplish their intercellular movement. A group of positive-strand RNA plant viruses requires three viral proteins (TGBp1, TGBp2, and TGBp3) that are encoded by an evolutionarily conserved genetic module of three partially overlapping open reading frames (ORFs), termed the triple gene block (TGB). However, how these three viral movement proteins function cooperatively in viral intercellular movement is still elusive. Using a novel in vivo double-stranded RNA (dsRNA) labeling system, we showed that the dsRNAs generated by potato virus X (PVX) RNA-dependent RNA polymerase (RdRp) are colocalized with viral RdRp, which are further tightly covered by "chain mail"-like TGBp2 aggregates and localizes alongside TGBp3 aggregates. We also discovered that TGBp2 interacts with the C-terminal domain of PVX RdRp, and this interaction is required for the localization of TGBp3 and itself to the RdRp/dsRNA bodies. Moreover, we reveal that the central and C-terminal hydrophilic domains of TGBp2 are required to interact with viral RdRp. Finally, we demonstrate that knockout of the entire TGBp2 or the domain involved in interacting with viral RdRp attenuates both PVX replication and movement. Collectively, these findings suggest that TGBp2 plays dual functional roles in PVX replication and intercellular movement.IMPORTANCE Many plant viruses contain three partially overlapping open reading frames (ORFs), termed the triple gene block (TGB), for intercellular movement. However, how the corresponding three proteins coordinate their functions remains obscure. In the present study, we provided multiple lines of evidence supporting the notion that PVX TGBp2 functions as the molecular adaptor bridging the interaction between the RdRp/dsRNA body and TGBp3 by forming "chain mail"-like structures in the RdRp/dsRNA body, which can also enhance viral replication. Taken together, our results provide new insights into the replication and movement of PVX and possibly also other TGB-containing plant viruses.
Keywords: Potato virus X; TGB; TGBp2; movement; replication.
Copyright © 2019 American Society for Microbiology.
Figures





Similar articles
-
The potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell-to-cell movement.Virology. 2003 Jul 20;312(1):35-48. doi: 10.1016/s0042-6822(03)00180-6. Virology. 2003. PMID: 12890619
-
Subcellular targeting and interactions among the Potato virus X TGB proteins.Virology. 2007 Oct 25;367(2):375-89. doi: 10.1016/j.virol.2007.05.022. Epub 2007 Jul 5. Virology. 2007. PMID: 17610926
-
The cysteine residues at the C-terminal tail of Bamboo mosaic virus triple gene block protein 2 are critical for efficient plasmodesmata localization of protein 1 in the same block.Virology. 2017 Jan 15;501:47-53. doi: 10.1016/j.virol.2016.11.005. Epub 2016 Nov 15. Virology. 2017. PMID: 27863274
-
Molecular biology of potexviruses: recent advances.J Gen Virol. 2007 Jun;88(Pt 6):1643-1655. doi: 10.1099/vir.0.82667-0. J Gen Virol. 2007. PMID: 17485523 Review.
-
A new cell-to-cell transport model for Potexviruses.Mol Plant Microbe Interact. 2005 Apr;18(4):283-90. doi: 10.1094/MPMI-18-0283. Mol Plant Microbe Interact. 2005. PMID: 15828680 Review.
Cited by
-
Mechanisms of plant virus cell-to-cell transport: new lessons from complementation studies.Front Plant Sci. 2024 Sep 18;15:1453464. doi: 10.3389/fpls.2024.1453464. eCollection 2024. Front Plant Sci. 2024. PMID: 39359631 Free PMC article. No abstract available.
-
Alphaflexiviridae in Focus: Genomic Signatures, Conserved Elements and Viral-Driven Cellular Remodeling.Viruses. 2025 Apr 24;17(5):611. doi: 10.3390/v17050611. Viruses. 2025. PMID: 40431623 Free PMC article. Review.
-
The 6-kilodalton peptide 1 in plant viruses of the family Potyviridae is a viroporin.Proc Natl Acad Sci U S A. 2024 May 21;121(21):e2401748121. doi: 10.1073/pnas.2401748121. Epub 2024 May 13. Proc Natl Acad Sci U S A. 2024. PMID: 38739789 Free PMC article.
-
Translation initiation landscape profiling reveals hidden open-reading frames required for the pathogenesis of tomato yellow leaf curl Thailand virus.Plant Cell. 2022 Apr 26;34(5):1804-1821. doi: 10.1093/plcell/koac019. Plant Cell. 2022. PMID: 35080617 Free PMC article.
-
Imaging Techniques to Study Plant Virus Replication and Vertical Transmission.Viruses. 2021 Feb 25;13(3):358. doi: 10.3390/v13030358. Viruses. 2021. PMID: 33668729 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

