Involvement of a Nonstructural Protein in Poliovirus Capsid Assembly
- PMID: 30541849
- PMCID: PMC6384072
- DOI: 10.1128/JVI.01447-18
Involvement of a Nonstructural Protein in Poliovirus Capsid Assembly
Abstract
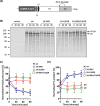
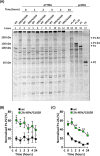
Virus capsid proteins must perform a number of roles. These include self-assembly and maintaining stability under challenging environmental conditions, while retaining the conformational flexibility necessary to uncoat and deliver the viral genome into a host cell. Fulfilling these roles could place conflicting constraints on the innate abilities encoded within the protein sequences. In a previous study, we identified a number of mutations within the capsid-coding sequence of poliovirus (PV) that were established in the population during selection for greater thermostability by sequential treatment at progressively higher temperatures. Two mutations in the VP1 protein acquired at an early stage were maintained throughout this selection procedure. One of these mutations prevented virion assembly when introduced into a wild-type (wt) infectious clone. Here we show, by sequencing beyond the capsid-coding region of the heat-selected virions, that two mutations had arisen within the coding region of the 2A protease. Both mutations were maintained throughout the selection process. Introduction of these mutations into a wt infectious clone by site-directed mutagenesis considerably reduced replication. However, they permitted a low level of assembly of infectious virions containing the otherwise lethal mutation in VP1. The 2Apro mutations were further shown to slow the kinetics of viral polyprotein processing, and we suggest that this delay improves the correct folding of the mutant capsid precursor protein to permit virion assembly.IMPORTANCE RNA viruses, including poliovirus, evolve rapidly due to the error-prone nature of the polymerase enzymes involved in genome replication. Fixation of advantageous mutations may require the acquisition of complementary mutations which can act in concert to achieve a favorable phenotype. This study highlights a compensatory role of a nonstructural regulatory protein, 2Apro, for an otherwise lethal mutation of the structural VP1 protein to facilitate increased thermal resistance. Studying how viruses respond to selection pressures is important for understanding mechanisms which underpin emergence of resistance and could be applied to the future development of antiviral agents and vaccines.
Keywords: 2Apro; cleavage; evolution; poliovirus; selection.
Copyright © 2019 Adeyemi et al.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

