Rotavirus Infection Alters Splicing of the Stress-Related Transcription Factor XBP1
- PMID: 30541862
- PMCID: PMC6384074
- DOI: 10.1128/JVI.01739-18
Rotavirus Infection Alters Splicing of the Stress-Related Transcription Factor XBP1
Abstract
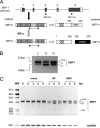
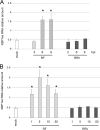
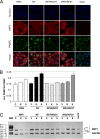
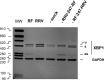
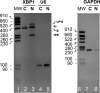
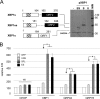
XBP1 is a stress-regulated transcription factor also involved in mammalian host defenses and innate immune response. Our investigation of XBP1 RNA splicing during rotavirus infection revealed that an additional XBP1 RNA (XBP1es) that corresponded to exon skipping in the XBP1 pre-RNA is induced depending on the rotavirus strain used. We show that the translation product of XBP1es (XBP1es) has trans-activation properties similar to those of XBP1 on ER stress response element (ERSE) containing promoters. Using monoreassortant between ES+ ("skipping") and ES- ("nonskipping") strains of rotavirus, we show that gene 7 encoding the viral translation enhancer NSP3 is involved in this phenomenon and that exon skipping parallels the nuclear relocalization of cytoplasmic PABP. We further show, using recombinant rotaviruses carrying chimeric gene 7, that the ES+ phenotype is linked to the eIF4G-binding domain of NSP3. Because the XBP1 transcription factor is involved in stress and immunological responses, our results suggest an alternative way to activate XBP1 upon viral infection or nuclear localization of PABP.IMPORTANCE Rotavirus is one of the most important pathogens causing severe gastroenteritis in young children worldwide. Here we show that infection with several rotavirus strains induces an alternative splicing of the RNA encoding the stressed-induced transcription factor XBP1. The genetic determinant of XBP1 splicing is the viral RNA translation enhancer NSP3. Since XBP1 is involved in cellular stress and immune responses and since the XBP1 protein made from the alternatively spliced RNA is an active transcription factor, our observations raise the question of whether alternative splicing is a cellular response to rotavirus infection.
Keywords: NSP3; PABP; XBP1; immune response; nuclear transport; rotavirus; splicing; stress response.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Rotavirus NSP3 Is a Translational Surrogate of the Poly(A) Binding Protein-Poly(A) Complex.J Virol. 2015 Sep;89(17):8773-82. doi: 10.1128/JVI.01402-15. Epub 2015 Jun 10. J Virol. 2015. PMID: 26063427 Free PMC article.
-
Species A rotavirus NSP3 acquires its translation inhibitory function prior to stable dimer formation.PLoS One. 2017 Jul 24;12(7):e0181871. doi: 10.1371/journal.pone.0181871. eCollection 2017. PLoS One. 2017. PMID: 28738064 Free PMC article.
-
Rotavirus variant replicates efficiently although encoding an aberrant NSP3 that fails to induce nuclear localization of poly(A)-binding protein.J Gen Virol. 2012 Jul;93(Pt 7):1483-1494. doi: 10.1099/vir.0.041830-0. Epub 2012 Mar 21. J Gen Virol. 2012. PMID: 22442114 Free PMC article.
-
Unconventional splicing of XBP-1 mRNA in the unfolded protein response.Antioxid Redox Signal. 2007 Dec;9(12):2323-33. doi: 10.1089/ars.2007.1800. Antioxid Redox Signal. 2007. PMID: 17979529 Review.
-
Rotavirus RNA replication and gene expression.Novartis Found Symp. 2001;238:64-77; discussion 77-81. doi: 10.1002/0470846534.ch5. Novartis Found Symp. 2001. PMID: 11444036 Review.
Cited by
-
Viral modulation of cellular RNA alternative splicing: A new key player in virus-host interactions?Wiley Interdiscip Rev RNA. 2019 Sep;10(5):e1543. doi: 10.1002/wrna.1543. Epub 2019 Apr 29. Wiley Interdiscip Rev RNA. 2019. PMID: 31034770 Free PMC article. Review.
-
ATF6-Mediated Unfolded Protein Response Facilitates Adeno-associated Virus 2 (AAV2) Transduction by Releasing the Suppression of the AAV Receptor on Endoplasmic Reticulum Stress.J Virol. 2022 Feb 9;96(3):e0110321. doi: 10.1128/JVI.01103-21. Epub 2021 Dec 1. J Virol. 2022. PMID: 34851146 Free PMC article.
-
XBP1 is required in Th2 polarization induction in airway allergy.Theranostics. 2022 Jul 11;12(12):5337-5349. doi: 10.7150/thno.75100. eCollection 2022. Theranostics. 2022. PMID: 35910793 Free PMC article.
-
Treading a HOSTile path: Mapping the dynamic landscape of host cell-rotavirus interactions to explore novel host-directed curative dimensions.Virulence. 2021 Dec;12(1):1022-1062. doi: 10.1080/21505594.2021.1903198. Virulence. 2021. PMID: 33818275 Free PMC article. Review.
-
Localization and Functional Roles of Components of the Translation Apparatus in the Eukaryotic Cell Nucleus.Cells. 2021 Nov 19;10(11):3239. doi: 10.3390/cells10113239. Cells. 2021. PMID: 34831461 Free PMC article. Review.
References
-
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, Martinez J. 2014. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J 33:2922–2936. doi:10.15252/embj.201490332. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

