The Interaction of Bluetongue Virus VP6 and Genomic RNA Is Essential for Genome Packaging
- PMID: 30541863
- PMCID: PMC6384066
- DOI: 10.1128/JVI.02023-18
The Interaction of Bluetongue Virus VP6 and Genomic RNA Is Essential for Genome Packaging
Abstract
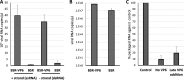
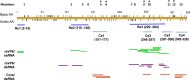
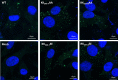
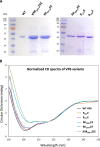
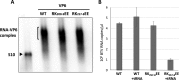
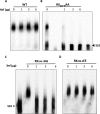
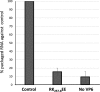
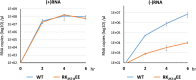
The genomes of the Reoviridae, including the animal pathogen bluetongue virus (BTV), are multisegmented double-stranded RNA (dsRNA). During replication, single-stranded (ss) positive-sense RNA segments are packaged into the assembling virus capsid, triggering genomic dsRNA synthesis. However, exactly how this packaging event occurs is not clear. A minor capsid protein, VP6, unique for the orbiviruses, has been proposed to be involved in the RNA-packaging process. In this study, we sought to characterize the RNA binding activity of VP6 and its functional relevance. A novel proteomic approach was utilized to map the ssRNA/dsRNA binding sites of a purified recombinant protein and the genomic dsRNA binding sites of the capsid-associated VP6. The data revealed that each VP6 protein has multiple distinct RNA-binding regions and that only one region is shared between recombinant and capsid-associated VP6. A combination of targeted mutagenesis and reverse genetics identified the RNA-binding region that is essential for virus replication. Using an in vitro RNA-binding competition assay, a unique cell-free assembly assay, and an in vivo single-cycle replication assay, it was possible to identify a motif within the shared binding region that binds BTV ssRNA preferentially in a manner consistent with specific RNA recruitment during capsid assembly. These data highlight the critical roles that this unique protein plays in orbivirus genome packaging and replication.IMPORTANCE Genome packaging is a critical stage during virus replication. For viruses with segmented genomes, the genome segments need to be correctly packaged into a newly formed capsid. However, the detailed mechanism of this packaging is unclear. Here we focus on VP6, a minor viral protein of bluetongue virus, which is critical for genome packaging. We used multiple approaches, including a robust RNA-protein fingerprinting assay, to map the ssRNA binding sites of recombinant VP6 and the genomic dsRNA binding sites of capsid-associated VP6. By these means, together with virological and biochemical methods, we identify the viral RNA-packaging motif of a segmented dsRNA virus for the first time.
Keywords: RNA-protein interaction; double-stranded RNA virus; genome packaging.
Copyright © 2019 Sung et al.
Figures

Similar articles
-
RNA Origami: Packaging a Segmented Genome in Orbivirus Assembly and Replication.Viruses. 2021 Sep 15;13(9):1841. doi: 10.3390/v13091841. Viruses. 2021. PMID: 34578422 Free PMC article. Review.
-
Bluetongue virus assembly and exit pathways.Adv Virus Res. 2020;108:249-273. doi: 10.1016/bs.aivir.2020.08.002. Epub 2020 Sep 16. Adv Virus Res. 2020. PMID: 33837718 Free PMC article. Review.
-
Interaction between a Unique Minor Protein and a Major Capsid Protein of Bluetongue Virus Controls Virus Infectivity.J Virol. 2018 Jan 17;92(3):e01784-17. doi: 10.1128/JVI.01784-17. Print 2018 Feb 1. J Virol. 2018. PMID: 29142128 Free PMC article.
-
RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.Cell. 2024 Apr 25;187(9):2236-2249.e17. doi: 10.1016/j.cell.2024.03.007. Epub 2024 Apr 12. Cell. 2024. PMID: 38614100 Free PMC article.
-
Disruption of Specific RNA-RNA Interactions in a Double-Stranded RNA Virus Inhibits Genome Packaging and Virus Infectivity.PLoS Pathog. 2015 Dec 8;11(12):e1005321. doi: 10.1371/journal.ppat.1005321. eCollection 2015 Dec. PLoS Pathog. 2015. PMID: 26646790 Free PMC article.
Cited by
-
RNA Origami: Packaging a Segmented Genome in Orbivirus Assembly and Replication.Viruses. 2021 Sep 15;13(9):1841. doi: 10.3390/v13091841. Viruses. 2021. PMID: 34578422 Free PMC article. Review.
-
In Vitro Reassortment between Endemic Bluetongue Viruses Features Global Shifts in Segment Frequencies and Preferred Segment Combinations.Microorganisms. 2021 Feb 16;9(2):405. doi: 10.3390/microorganisms9020405. Microorganisms. 2021. PMID: 33669284 Free PMC article.
-
Bluetongue virus assembly and exit pathways.Adv Virus Res. 2020;108:249-273. doi: 10.1016/bs.aivir.2020.08.002. Epub 2020 Sep 16. Adv Virus Res. 2020. PMID: 33837718 Free PMC article. Review.
-
Vaccination as a Strategy to Prevent Bluetongue Virus Vertical Transmission.Pathogens. 2021 Nov 22;10(11):1528. doi: 10.3390/pathogens10111528. Pathogens. 2021. PMID: 34832683 Free PMC article. Review.
-
A multidisciplinary approach to the identification of the protein-RNA connectome in double-stranded RNA virus capsids.Nucleic Acids Res. 2023 Jun 9;51(10):5210-5227. doi: 10.1093/nar/gkad274. Nucleic Acids Res. 2023. PMID: 37070191 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

