Inference of Large-scale Time-delayed Gene Regulatory Network with Parallel MapReduce Cloud Platform
- PMID: 30542062
- PMCID: PMC6290780
- DOI: 10.1038/s41598-018-36180-y
Inference of Large-scale Time-delayed Gene Regulatory Network with Parallel MapReduce Cloud Platform
Abstract
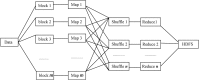
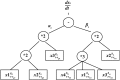
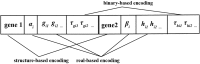
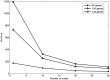
Inference of gene regulatory network (GRN) is crucial to understand intracellular physiological activity and function of biology. The identification of large-scale GRN has been a difficult and hot topic of system biology in recent years. In order to reduce the computation load for large-scale GRN identification, a parallel algorithm based on restricted gene expression programming (RGEP), namely MPRGEP, is proposed to infer instantaneous and time-delayed regulatory relationships between transcription factors and target genes. In MPRGEP, the structure and parameters of time-delayed S-system (TDSS) model are encoded into one chromosome. An original hybrid optimization approach based on genetic algorithm (GA) and gene expression programming (GEP) is proposed to optimize TDSS model with MapReduce framework. Time-delayed GRNs (TDGRN) with hundreds of genes are utilized to test the performance of MPRGEP. The experiment results reveal that MPRGEP could infer more accurately gene regulatory network than other state-of-art methods, and obtain the convincing speedup.
Conflict of interest statement
The authors declare no competing interests.
Figures








Similar articles
-
Designing a parallel evolutionary algorithm for inferring gene networks on the cloud computing environment.BMC Syst Biol. 2014 Jan 16;8:5. doi: 10.1186/1752-0509-8-5. BMC Syst Biol. 2014. PMID: 24428926 Free PMC article.
-
TDSDMI: Inference of time-delayed gene regulatory network using S-system model with delayed mutual information.Comput Biol Med. 2016 May 1;72:218-25. doi: 10.1016/j.compbiomed.2016.03.024. Epub 2016 Mar 30. Comput Biol Med. 2016. PMID: 27058285
-
A new asynchronous parallel algorithm for inferring large-scale gene regulatory networks.PLoS One. 2015 Mar 25;10(3):e0119294. doi: 10.1371/journal.pone.0119294. eCollection 2015. PLoS One. 2015. PMID: 25807392 Free PMC article.
-
Computational prediction of gene regulatory networks in plant growth and development.Curr Opin Plant Biol. 2019 Feb;47:96-105. doi: 10.1016/j.pbi.2018.10.005. Epub 2018 Nov 14. Curr Opin Plant Biol. 2019. PMID: 30445315 Review.
-
A Review of Computational Approach for S-system-based Modeling of Gene Regulatory Network.Methods Mol Biol. 2024;2719:133-152. doi: 10.1007/978-1-0716-3461-5_8. Methods Mol Biol. 2024. PMID: 37803116 Review.
Cited by
-
Inference of Molecular Regulatory Systems Using Statistical Path-Consistency Algorithm.Entropy (Basel). 2022 May 13;24(5):693. doi: 10.3390/e24050693. Entropy (Basel). 2022. PMID: 35626576 Free PMC article.
-
A non-linear reverse-engineering method for inferring genetic regulatory networks.PeerJ. 2020 Apr 29;8:e9065. doi: 10.7717/peerj.9065. eCollection 2020. PeerJ. 2020. PMID: 32391205 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

