The translation of non-canonical open reading frames controls mucosal immunity
- PMID: 30542152
- PMCID: PMC6939389
- DOI: 10.1038/s41586-018-0794-7
The translation of non-canonical open reading frames controls mucosal immunity
Abstract
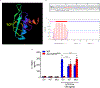
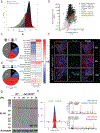
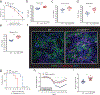
The annotation of the mammalian protein-coding genome is incomplete. Arbitrary size restriction of open reading frames (ORFs) and the absolute requirement for a methionine codon as the sole initiator of translation have constrained the identification of potentially important transcripts with non-canonical protein-coding potential1,2. Here, using unbiased transcriptomic approaches in macrophages that respond to bacterial infection, we show that ribosomes associate with a large number of RNAs that were previously annotated as 'non-protein coding'. Although the idea that such non-canonical ORFs can encode functional proteins is controversial3,4, we identify a range of short and non-ATG-initiated ORFs that can generate stable and spatially distinct proteins. Notably, we show that the translation of a new ORF 'hidden' within the long non-coding RNA Aw112010 is essential for the orchestration of mucosal immunity during both bacterial infection and colitis. This work expands our interpretation of the protein-coding genome and demonstrates that proteinaceous products generated from non-canonical ORFs are crucial for the immune response in vivo. We therefore propose that the misannotation of non-canonical ORF-containing genes as non-coding RNAs may obscure the essential role of a multitude of previously undiscovered protein-coding genes in immunity and disease.
Figures










References
-
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI & Zamecnik PC A SOLUBLE RIBONUCLEIC ACID INTERMEDIATE IN PROTEIN SYNTHESIS. Journal of Biological Chemistry 231, 241–257 (1958). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

