Long non-coding RNA LINC01503 promotes colorectal cancer cell proliferation and invasion by regulating miR-4492/FOXK1 signaling
- PMID: 30542444
- PMCID: PMC6257603
- DOI: 10.3892/etm.2018.6775
Long non-coding RNA LINC01503 promotes colorectal cancer cell proliferation and invasion by regulating miR-4492/FOXK1 signaling
Abstract
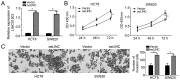
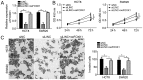
Increasing evidence indicates that long non-coding RNAs (lncRNAs) are closely associated with the progression of human cancer, including colorectal cancer (CRC). A previous study suggested that lncRNA LINC01503 promotes squamous cell carcinoma progression. However, the function of LINC01503 in CRC has remained elusive. The present study indicated that LINC01503 was significantly upregulated in CRC tissues compared with that in adjacent normal tissues as detected by reverse transcription-quantitative polymerase chain reaction. It was demonstrated that knockdown of long intergenic non-protein coding RNA (LINC)01503 markedly inhibited the proliferation and invasion of CRC cells, whereas overexpression of LINC01503 had the opposite effects, as indicated by Cell Counting kit-8 and Transwell assays. Mechanistically, it was revealed that LINC01503 serves as a sponge for microRNA (miR)-4492, which targets forkhead box K1 (FOXK1) in CRC cells. In addition, luciferase reporter assays demonstrated the direct binding of miR-4492 mimics to LINC01503 and to a sequence in the 3'-untranslated region of FOXK1. Furthermore, it was demonstrated that overexpression of LINC01503 reduced the availability of miR-4492 in CRC cells. Furthermore, miR-4492 mimics inhibited FOXK1 expression, while simultaneous overexpression of LINC01503 abolished this effect. Finally, it was demonstrated that restoration of FOXK1 abolished the inhibitory effect of LINC01503 knockdown on CRC cell proliferation and invasion. Taken together, the present results suggested that LINC01503 promotes CRC progression via acting as a competing endogenous RNA for miR-4492/FOXK1.
Keywords: LINC01503; colorectal cancer; forkhead box K1; invasion; microRNA-4492; proliferation.
Figures



Similar articles
-
Transcription factor forkhead box K1 regulates miR-32 expression and enhances cell proliferation in colorectal cancer.Oncol Lett. 2021 May;21(5):407. doi: 10.3892/ol.2021.12668. Epub 2021 Mar 22. Oncol Lett. 2021. PMID: 33841568 Free PMC article.
-
The lncRNA XIST promotes colorectal cancer cell growth through regulating the miR-497-5p/FOXK1 axis.Cancer Cell Int. 2020 Dec 10;20(1):553. doi: 10.1186/s12935-020-01647-4. Cancer Cell Int. 2020. Retraction in: Cancer Cell Int. 2021 Sep 14;21(1):484. doi: 10.1186/s12935-021-02205-2. PMID: 33298041 Free PMC article. Retracted.
-
Long non-coding RNA SNHG12 promotes proliferation and invasion of colorectal cancer cells by acting as a molecular sponge of microRNA-16.Exp Ther Med. 2019 Aug;18(2):1212-1220. doi: 10.3892/etm.2019.7650. Epub 2019 Jun 7. Exp Ther Med. 2019. PMID: 31316616 Free PMC article.
-
Roles, underlying mechanisms and clinical significances of LINC01503 in human cancers.Pathol Res Pract. 2024 Feb;254:155125. doi: 10.1016/j.prp.2024.155125. Epub 2024 Jan 17. Pathol Res Pract. 2024. PMID: 38241778 Review.
-
The Function of FoxK Transcription Factors in Diseases.Front Physiol. 2022 Jul 12;13:928625. doi: 10.3389/fphys.2022.928625. eCollection 2022. Front Physiol. 2022. PMID: 35903069 Free PMC article. Review.
Cited by
-
Long intergenic noncoding RNA smad7 (Linc-smad7) promotes the epithelial-mesenchymal transition of HCC by targeting the miR-125b/SIRT6 axis.Cancer Med. 2020 Dec;9(23):9123-9137. doi: 10.1002/cam4.3515. Epub 2020 Oct 10. Cancer Med. 2020. PMID: 33037850 Free PMC article.
-
LncRNA SNHG7 promotes the proliferation of nasopharyngeal carcinoma by miR-514a-5p/ELAVL1 axis.BMC Cancer. 2020 May 5;20(1):376. doi: 10.1186/s12885-020-06775-8. BMC Cancer. 2020. PMID: 32370736 Free PMC article.
-
Long Non-coding RNA LINC01503 Promotes Gastric Cardia Adenocarcinoma Progression via miR-133a-5p/VIM Axis and EMT Process.Dig Dis Sci. 2021 Oct;66(10):3391-3403. doi: 10.1007/s10620-020-06690-9. Epub 2020 Nov 17. Dig Dis Sci. 2021. PMID: 33200343
-
Long Non-Coding RNA Signatures Associated with Ferroptosis Predict Prognosis in Colorectal Cancer.Int J Gen Med. 2022 Jan 4;15:33-43. doi: 10.2147/IJGM.S331378. eCollection 2022. Int J Gen Med. 2022. PMID: 35018112 Free PMC article.
-
Integrative analysis of ceRNA network reveals functional lncRNAs associated with independent recurrent prognosis in colon adenocarcinoma.Cancer Cell Int. 2021 Jul 5;21(1):352. doi: 10.1186/s12935-021-02069-6. Cancer Cell Int. 2021. PMID: 34225739 Free PMC article.
References
LinkOut - more resources
Full Text Sources
