Bioinformatics analysis reveals different gene expression patterns in the annulus fibrosis and nucleus pulpous during intervertebral disc degeneration
- PMID: 30542457
- PMCID: PMC6257805
- DOI: 10.3892/etm.2018.6884
Bioinformatics analysis reveals different gene expression patterns in the annulus fibrosis and nucleus pulpous during intervertebral disc degeneration
Abstract
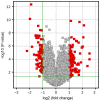
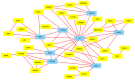
Degeneration of the intervertebral disc (IVD), which consists of the annulus fibrosus (AF) and nucleus pulposus (NP), is a multifactorial physiological process associated with lower back pain. Despite decades of research, the knowledge of the underlying molecular mechanisms of IVD degeneration (IDD) has remained limited. The present study aimed to reveal the differential gene expression patterns in AF and NP during the process of IDD and to identify key biomarkers contributing to these differences. The microarray dataset GSE70362 containing 24 AF and 24 NP samples was retrieved from the Gene Expression Omnibus database. Of these, 8 healthy samples were discarded. GeneSpring11.5 software was employed to identify differentially expressed genes (DEGs). Metascape online tools were used to perform enrichment analyses. Finally, the DEGs were mapped with the Search Tool for the Retrieval of Interacting Genes, and a protein-protein interaction (PPI) network was constructed in Cytoscape software. A total of 87 DEGs were identified. Gene ontology enrichment revealed that these DEGs were mainly involved in the inflammatory response, the extracellular matrix and RNA polymerase II transcription factor activity. Pathway enrichment revealed that the DEGs were mainly involved in the transforming growth factor (TGF-β) and estrogen signaling pathways. Matrix metalloproteinase (MMP)1 and interleukin (IL)6 were included in the genes enriched in rheumatoid arthritis, whereas bone morphogenetic protein (BMP)2 and thrombospondin 1 (THBS1) were among the genes enriched in the TGF-β signaling pathway. In the PPI network, IL6 was identified as the central gene. In conclusion, as MMP1 has been demonstrated degrade collagen III at higher rates compared with other types of collagen (which is at a higher quantity in AF than NP), collagen types may be in different distribution patterns, which may contribute to the upregulation of MMP1 in AF. Differences in the expression of BMP2, ESR1 and THBS1 may explain for the pathological differences between AF and NP. IL6 may have a key role in different degeneration processes in AF and NP.
Keywords: differential expression; enrichment analysis; protein-protein interaction network.
Figures




Similar articles
-
Identification of Key Genes Potentially Related to Intervertebral Disk Degeneration by Microarray Analysis.Genet Test Mol Biomarkers. 2019 Sep;23(9):610-617. doi: 10.1089/gtmb.2019.0043. Epub 2019 Aug 1. Genet Test Mol Biomarkers. 2019. PMID: 31368816
-
Gene expression profile identifies potential biomarkers for human intervertebral disc degeneration.Mol Med Rep. 2017 Dec;16(6):8665-8672. doi: 10.3892/mmr.2017.7741. Epub 2017 Oct 9. Mol Med Rep. 2017. PMID: 29039500 Free PMC article.
-
Identification of key potential targets for TNF-α/TNFR1-related intervertebral disc degeneration by bioinformatics analysis.Connect Tissue Res. 2021 Sep;62(5):531-541. doi: 10.1080/03008207.2020.1797709. Epub 2020 Aug 26. Connect Tissue Res. 2021. PMID: 32686499
-
Biomechanics in Annulus Fibrosus Degeneration and Regeneration.Adv Exp Med Biol. 2018;1078:409-420. doi: 10.1007/978-981-13-0950-2_21. Adv Exp Med Biol. 2018. PMID: 30357635 Review.
-
Intervertebral disc development and disease-related genetic polymorphisms.Genes Dis. 2016 Apr 23;3(3):171-177. doi: 10.1016/j.gendis.2016.04.006. eCollection 2016 Sep. Genes Dis. 2016. PMID: 30258887 Free PMC article. Review.
Cited by
-
Microarray analysis reveals an inflammatory transcriptomic signature in peripheral blood for sciatica.BMC Neurol. 2021 Feb 3;21(1):50. doi: 10.1186/s12883-021-02078-y. BMC Neurol. 2021. PMID: 33535986 Free PMC article.
-
Integrated traditional Chinese medicine alleviates sciatica while regulating gene expression in peripheral blood.J Orthop Surg Res. 2021 Feb 11;16(1):130. doi: 10.1186/s13018-021-02280-1. J Orthop Surg Res. 2021. PMID: 33573686 Free PMC article.
-
Single-Cell RNA-Seq Analysis of Cells from Degenerating and Non-Degenerating Intervertebral Discs from the Same Individual Reveals New Biomarkers for Intervertebral Disc Degeneration.Int J Mol Sci. 2022 Apr 3;23(7):3993. doi: 10.3390/ijms23073993. Int J Mol Sci. 2022. PMID: 35409356 Free PMC article.
-
FOXO3-Activated circFGFBP1 Inhibits Extracellular Matrix Degradation and Nucleus Pulposus Cell Death via miR-9-5p/BMP2 Axis in Intervertebral Disc Degeneration In Vivo and In Vitro.Pharmaceuticals (Basel). 2023 Mar 22;16(3):473. doi: 10.3390/ph16030473. Pharmaceuticals (Basel). 2023. PMID: 36986573 Free PMC article.
-
Integrated weighted gene coexpression network analysis identifies Frizzled 2 (FZD2) as a key gene in invasive malignant pleomorphic adenoma.J Transl Med. 2022 Jan 5;20(1):15. doi: 10.1186/s12967-021-03204-7. J Transl Med. 2022. PMID: 34986855 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
