Transcriptional profiling analysis predicts potential biomarkers for glaucoma: HGF, AKR1B10 and AKR1C3
- PMID: 30542465
- PMCID: PMC6257105
- DOI: 10.3892/etm.2018.6875
Transcriptional profiling analysis predicts potential biomarkers for glaucoma: HGF, AKR1B10 and AKR1C3
Abstract
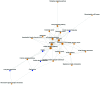
Glaucoma results in damage to the optic nerve and vision loss. The aim of this study was to screen more accurate biomarkers and targets for glaucoma. The datasets E-GEOD-7144 and E-MEXP-3427 were screened for differently expressed genes (DEGs) by significance analysis of microarrays. Functional and pathway enrichment analysis were processed. Pathway relationship networks and gene co-expression networks were constructed. DEGs of disease and treatment with the same symbols were of interest. RT-qPCR was processed to verify the expression of key DEGs. A total of 1,019 DEGs of glaucoma were identified and 93 DEGs in transforming growth factor-β1 (TGF-β1) and TGF-β1-2 treatment cases compared with the normal control group. Pathway relationship network of glaucoma was constructed with 25 nodes. The pathway relationship network of TGF-β1 and -2 treatment groups was constructed with 11 nodes. Glaucoma-related DEGs in GO terms and pathways were inserted and 180 common DEGs were obtained. Then, gene co-expression network of glaucoma-related DEGs was constructed with 91 nodes. Furthermore, DEGs of TGF-β1 and -2 treated glaucoma in GO terms and pathways were inserted, and 29 common DEGs were identified. Based on these DEGs, gene co-expression network was constructed with 12 nodes and 16 edges. Finally, a total of 6 important DEGs of disease and treatment were inserted and obtained. They were HGF, AKR1B10, AKR1C3, PPAP2B, INHBA and BCAT1. The expression of HGF, AKR1B10 and AKR1C3 was decreased in glaucoma samples and treatment samples. In conclusion, HGF, AKR1B10 and AKR1C3 may be key genes for glaucoma diagnosis and treatment.
Keywords: TGF-β1 and −2; biomarker; glaucoma; pathway.
Figures



Similar articles
-
Transcriptome profiling analysis reveals biomarkers in colon cancer samples of various differentiation.Oncol Lett. 2018 Jul;16(1):48-54. doi: 10.3892/ol.2018.8668. Epub 2018 May 8. Oncol Lett. 2018. PMID: 29928385 Free PMC article.
-
Analysis of differentially expressed genes in white blood cells isolated from patients with major burn injuries.Exp Ther Med. 2017 Oct;14(4):2931-2936. doi: 10.3892/etm.2017.4899. Epub 2017 Aug 7. Exp Ther Med. 2017. PMID: 28966676 Free PMC article.
-
Bioinformatics analysis to identify the differentially expressed genes of glaucoma.Mol Med Rep. 2015 Oct;12(4):4829-36. doi: 10.3892/mmr.2015.4030. Epub 2015 Jul 2. Mol Med Rep. 2015. PMID: 26135629 Free PMC article.
-
Smoking and drinking influence the advancing of ischemic stroke disease by targeting PTGS2 and TNFAIP3.Exp Ther Med. 2018 Jul;16(1):61-66. doi: 10.3892/etm.2018.6138. Epub 2018 May 7. Exp Ther Med. 2018. PMID: 29977356 Free PMC article.
-
Series test of cluster and network analysis for lupus nephritis, before and after IFN-K-immunosuppressive therapy.Nephrology (Carlton). 2018 Nov;23(11):997-1006. doi: 10.1111/nep.13159. Nephrology (Carlton). 2018. PMID: 28869321
Cited by
-
AKR1B10 inhibits the proliferation and migration of gastric cancer via regulating epithelial-mesenchymal transition.Aging (Albany NY). 2021 Sep 22;13(18):22298-22314. doi: 10.18632/aging.203538. Epub 2021 Sep 22. Aging (Albany NY). 2021. PMID: 34552036 Free PMC article.
-
Updates on Genes and Genetic Mechanisms Implicated in Primary Angle-Closure Glaucoma.Appl Clin Genet. 2021 Mar 9;14:89-112. doi: 10.2147/TACG.S274884. eCollection 2021. Appl Clin Genet. 2021. PMID: 33727852 Free PMC article. Review.
-
Candidate SNP Markers Significantly Altering the Affinity of the TATA-Binding Protein for the Promoters of Human Genes Associated with Primary Open-Angle Glaucoma.Int J Mol Sci. 2024 Nov 28;25(23):12802. doi: 10.3390/ijms252312802. Int J Mol Sci. 2024. PMID: 39684516 Free PMC article.
References
LinkOut - more resources
Full Text Sources
