Design, synthesis, and anticancer activity evaluation of irreversible allosteric inhibitors of the ubiquitin-conjugating enzyme Ube2g2
- PMID: 30542531
- PMCID: PMC6238722
- DOI: 10.1039/c8md00320c
Design, synthesis, and anticancer activity evaluation of irreversible allosteric inhibitors of the ubiquitin-conjugating enzyme Ube2g2
Abstract
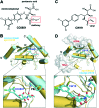
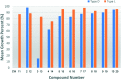
The RING finger-dependent ubiquitin ligase (E3) gp78, known as the tumor autocrine motility factor receptor, contributes to tumor progression. The protein interacts with its cognate ubiquitin-conjugating enzyme (E2), Ube2g2, via its RING domain and a unique region called G2BR that strongly binds to E2. The binding of G2BR to Ube2g2 allosterically enhances the binding of RING to E2, and the binding of RING triggers the departure of G2BR from E2 also in an allosteric fashion. Targeting these allosteric events, we developed a family of inhibitors that irreversibly block E2-E3 interactions and thereby eliminate the tumorigenic effect of gp78. One among 19 compounds screened with the NCI 60 tumor cell lines exhibited outstanding anticancer activities. At 10 μM, it caused >50% growth inhibition to 40% of the cell lines; at 100 μM, it showed lethiferous activity against most cell lines.
Figures



Similar articles
-
Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.EMBO J. 2013 Sep 11;32(18):2504-16. doi: 10.1038/emboj.2013.174. Epub 2013 Aug 13. EMBO J. 2013. PMID: 23942235 Free PMC article.
-
Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78.Mol Cell. 2009 Jun 26;34(6):674-85. doi: 10.1016/j.molcel.2009.05.010. Mol Cell. 2009. PMID: 19560420 Free PMC article.
-
Conformational Dynamics and Allostery in E2:E3 Interactions Drive Ubiquitination: gp78 and Ube2g2.Structure. 2017 May 2;25(5):794-805.e5. doi: 10.1016/j.str.2017.03.016. Epub 2017 Apr 20. Structure. 2017. PMID: 28434917 Free PMC article.
-
A nanobody that binds to the backside of the ubiquitin conjugating enzyme Ube2G2 differentially affects interactions with its partner E3 Ligases.Commun Biol. 2025 Apr 16;8(1):614. doi: 10.1038/s42003-025-08038-3. Commun Biol. 2025. PMID: 40234692 Free PMC article.
-
Ubiquitin-conjugating enzyme E2C: a potential cancer biomarker.Int J Biochem Cell Biol. 2014 Feb;47:113-7. doi: 10.1016/j.biocel.2013.11.023. Epub 2013 Dec 17. Int J Biochem Cell Biol. 2014. PMID: 24361302 Review.
Cited by
-
A Novel Confocal Scanning Protein-Protein Interaction Assay (PPI-CONA) Reveals Exceptional Selectivity and Specificity of CC0651, a Small Molecule Binding Enhancer of the Weak Interaction between the E2 Ubiquitin-Conjugating Enzyme CDC34A and Ubiquitin.Bioconjug Chem. 2024 Sep 18;35(9):1441-1449. doi: 10.1021/acs.bioconjchem.4c00345. Epub 2024 Aug 21. Bioconjug Chem. 2024. PMID: 39167708 Free PMC article.
-
The Molecular Basis of Ubiquitin-Conjugating Enzymes (E2s) as a Potential Target for Cancer Therapy.Int J Mol Sci. 2021 Mar 26;22(7):3440. doi: 10.3390/ijms22073440. Int J Mol Sci. 2021. PMID: 33810518 Free PMC article. Review.
-
Ubiquitination-Related miRNA-mRNA Interaction Is a Potential Mechanism in the Progression of Retinoblastoma.Invest Ophthalmol Vis Sci. 2021 Aug 2;62(10):3. doi: 10.1167/iovs.62.10.3. Invest Ophthalmol Vis Sci. 2021. PMID: 34347012 Free PMC article.
References
-
- Ceccarelli D. F., Tang X., Pelletier B., Orlicky S., Xie W., Plantevin V., Neculai D., Chou Y. C., Ogunjimi A., Al-Hakim A., Varelas X., Koszela J., Wasney G. A., Vedadi M., Dhe-Paganon S., Cox S., Xu S., Lopez-Girona A., Mercurio F., Wrana J., Durocher D., Meloche S., Webb D. R., Tyers M., Sicheri F. Cell. 2011;145:1075–1087. - PubMed
LinkOut - more resources
Full Text Sources

