Multiple evolutionary origins of giant viruses
- PMID: 30542614
- PMCID: PMC6259494
- DOI: 10.12688/f1000research.16248.1
Multiple evolutionary origins of giant viruses
Abstract
The nucleocytoplasmic large DNA viruses (NCLDVs) are a monophyletic group of diverse eukaryotic viruses that reproduce primarily in the cytoplasm of the infected cells and include the largest viruses currently known: the giant mimiviruses, pandoraviruses, and pithoviruses. With virions measuring up to 1.5 μm and genomes of up to 2.5 Mb, the giant viruses break the now-outdated definition of a virus and extend deep into the genome size range typical of bacteria and archaea. Additionally, giant viruses encode multiple proteins that are universal among cellular life forms, particularly components of the translation system, the signature cellular molecular machinery. These findings triggered hypotheses on the origin of giant viruses from cells, likely of an extinct fourth domain of cellular life, via reductive evolution. However, phylogenomic analyses reveal a different picture, namely multiple origins of giant viruses from smaller NCLDVs via acquisition of multiple genes from the eukaryotic hosts and bacteria, along with gene duplication. Thus, with regard to their origin, the giant viruses do not appear to qualitatively differ from the rest of the virosphere. However, the evolutionary forces that led to the emergence of virus gigantism remain enigmatic.
Keywords: gene gain; gene loss; giant viruses; phagocytosis; virus evolution; virus-host interaction.
Conflict of interest statement
No competing interests were disclosed.No competing interests were disclosed.No competing interests were disclosed.No competing interests were disclosed.No competing interests were disclosed.
Figures
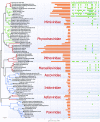
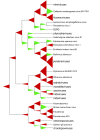
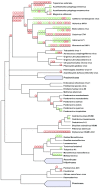
References
-
- Ivanowski D: Über die Mosaikkrankheit der Tabakspflanze. Bulletin Scientifique publié par l'Académie Impériale des Sciences de Saint-Pétersbourg / Nouvelle Serie III. 1892;35:67–70. Reference Source
-
- Beijerinck MW: Über ein Contagium vivum fluidum als Ursache der Fleckenkrankheit der Tabaksblätter. Verhandelingen der Koninklijke akademie van Wetenschappen te Amsterdam.1898;65:1–22. Reference Source
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

