Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence
- PMID: 30543681
- PMCID: PMC6307818
- DOI: 10.1371/journal.pgen.1007845
Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence
Abstract
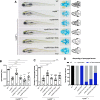
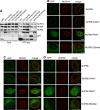
Nucleoporins build the nuclear pore complex (NPC), which, as sole gate for nuclear-cytoplasmic exchange, is of outmost importance for normal cell function. Defects in the process of nucleocytoplasmic transport or in its machinery have been frequently described in human diseases, such as cancer and neurodegenerative disorders, but only in a few cases of developmental disorders. Here we report biallelic mutations in the nucleoporin NUP88 as a novel cause of lethal fetal akinesia deformation sequence (FADS) in two families. FADS comprises a spectrum of clinically and genetically heterogeneous disorders with congenital malformations related to impaired fetal movement. We show that genetic disruption of nup88 in zebrafish results in pleiotropic developmental defects reminiscent of those seen in affected human fetuses, including locomotor defects as well as defects at neuromuscular junctions. Phenotypic alterations become visible at distinct developmental stages, both in affected human fetuses and in zebrafish, whereas early stages of development are apparently normal. The zebrafish phenotypes caused by nup88 deficiency are rescued by expressing wild-type Nup88 but not the disease-linked mutant forms of Nup88. Furthermore, using human and mouse cell lines as well as immunohistochemistry on fetal muscle tissue, we demonstrate that NUP88 depletion affects rapsyn, a key regulator of the muscle nicotinic acetylcholine receptor at the neuromuscular junction. Together, our studies provide the first characterization of NUP88 in vertebrate development, expand our understanding of the molecular events causing FADS, and suggest that variants in NUP88 should be investigated in cases of FADS.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, et al. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. Embo J. 1997;16(4):807–16. Epub 1997/02/17. 10.1093/emboj/16.4.807 ; PubMed Central PMCID: PMC1169681. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

