Septins Recognize and Entrap Dividing Bacterial Cells for Delivery to Lysosomes
- PMID: 30543779
- PMCID: PMC6299245
- DOI: 10.1016/j.chom.2018.11.005
Septins Recognize and Entrap Dividing Bacterial Cells for Delivery to Lysosomes
Abstract
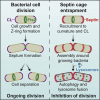
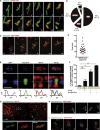
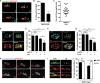
The cytoskeleton occupies a central role in cellular immunity by promoting bacterial sensing and antibacterial functions. Septins are cytoskeletal proteins implicated in various cellular processes, including cell division. Septins also assemble into cage-like structures that entrap cytosolic Shigella, yet how septins recognize bacteria is poorly understood. Here, we discover that septins are recruited to regions of micron-scale membrane curvature upon invasion and division by a variety of bacterial species. Cardiolipin, a curvature-specific phospholipid, promotes septin recruitment to highly curved membranes of Shigella, and bacterial mutants lacking cardiolipin exhibit less septin cage entrapment. Chemically inhibiting cell separation to prolong membrane curvature or reducing Shigella cell growth respectively increases and decreases septin cage formation. Once formed, septin cages inhibit Shigella cell division upon recruitment of autophagic and lysosomal machinery. Thus, recognition of dividing bacterial cells by the septin cytoskeleton is a powerful mechanism to restrict the proliferation of intracellular bacterial pathogens.
Keywords: FtsZ; Shigella; cardiolipin; cytoskeleton; membrane curvature; septins.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
Bacterial cell division is recognized by the septin cytoskeleton for restriction by autophagy.Autophagy. 2019 May;15(5):937-939. doi: 10.1080/15548627.2019.1586499. Epub 2019 Mar 11. Autophagy. 2019. PMID: 30857451 Free PMC article.
References
-
- Bi E., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. - PubMed
-
- Cherepanov P.P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

