Mechanisms of DNA Methyltransferase Recruitment in Mammals
- PMID: 30544749
- PMCID: PMC6316769
- DOI: 10.3390/genes9120617
Mechanisms of DNA Methyltransferase Recruitment in Mammals
Abstract
DNA methylation is an essential epigenetic mark in mammals. The proper distribution of this mark depends on accurate deposition and maintenance mechanisms, and underpins its functional role. This, in turn, depends on the precise recruitment and activation of de novo and maintenance DNA methyltransferases (DNMTs). In this review, we discuss mechanisms of recruitment of DNMTs by transcription factors and chromatin modifiers-and by RNA-and place these mechanisms in the context of biologically meaningful epigenetic events. We present hypotheses and speculations for future research, and underline the fundamental and practical benefits of better understanding the mechanisms that govern the recruitment of DNMTs.
Keywords: DNA methylation; DNA methyltransferases; epigenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

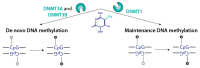



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

