Actinobaculum massiliense Proteome Profiled in Polymicrobial Urethral Catheter Biofilms
- PMID: 30544882
- PMCID: PMC6314084
- DOI: 10.3390/proteomes6040052
Actinobaculum massiliense Proteome Profiled in Polymicrobial Urethral Catheter Biofilms
Abstract
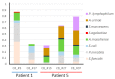
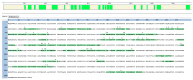
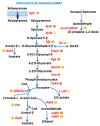
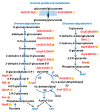
Actinobaculum massiliense, a Gram-positive anaerobic coccoid rod colonizing the human urinary tract, belongs to the taxonomic class of Actinobacteria. We identified A. massiliense as a cohabitant of urethral catheter biofilms (CB). The CBs also harbored more common uropathogens, such as Proteus mirabilis and Aerococcus urinae, supporting the notion that A. massiliense is adapted to a life style in polymicrobial biofilms. We isolated a clinical strain from a blood agar colony and used 16S rRNA gene sequencing and shotgun proteomics to confirm its identity as A. massiliense. We characterized this species by quantitatively comparing the bacterial proteome derived from in vitro growth with that of four clinical samples. The functional relevance of proteins with emphasis on nutrient import and the response to hostile host conditions, showing evidence of neutrophil infiltration, was analyzed. Two putative subtilisin-like proteases and a heme/oligopeptide transporter were abundant in vivo and are likely important for survival and fitness in the biofilm. Proteins facilitating uptake of xylose/glucuronate and oligopeptides, also highly expressed in vivo, may feed metabolites into mixed acid fermentation and peptidolysis pathways, respectively, to generate energy. A polyketide synthase predicted to generate a secondary metabolite that interacts with either the human host or co-colonizing microbes was also identified. The product of the PKS enzyme may contribute to A. massiliense fitness and persistence in the CBs.
Keywords: CAUTI; Keywords: actinobaculum; biofilm; catheter; host-pathogen interaction; metabolism; polymicrobial; proteome; urinary tract infection; uropathogen.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

References
-
- Lawson P.A., Falsen E., Akervall E., Vandamme P., Collins M.D. Characterization of some actinomyces-like isolates from human clinical specimens: Reclassification of Actinomyces suis (soltys and spratling) as Actinobaculum suis comb. Nov. And description of Actinobaculum schaalii sp. Nov. Int. J. Syst. Bacteriol. 1997;47:899–903. doi: 10.1099/00207713-47-3-899. - DOI - PubMed
-
- Reinhard M., Prag J., Kemp M., Andresen K., Klemmensen B., Hojlyng N., Sorensen S.H., Christensen J.J. Ten cases of Actinobaculum schaalii infection: Clinical relevance, bacterial identification, and antibiotic susceptibility. J. Clin. Microbiol. 2005;43:5305–5308. doi: 10.1128/JCM.43.10.5305-5308.2005. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

