Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation
- PMID: 30544982
- PMCID: PMC6316889
- DOI: 10.3390/genes9120620
Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation
Abstract
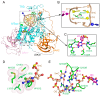
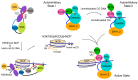
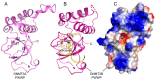
DNA methylation, one of the major epigenetic mechanisms, plays critical roles in regulating gene expression, genomic stability and cell lineage commitment. The establishment and maintenance of DNA methylation in mammals is achieved by two groups of DNA methyltransferases (DNMTs): DNMT3A and DNMT3B, which are responsible for installing DNA methylation patterns during gametogenesis and early embryogenesis, and DNMT1, which is essential for propagating DNA methylation patterns during replication. Both groups of DNMTs are multi-domain proteins, containing a large N-terminal regulatory region in addition to the C-terminal methyltransferase domain. Recent structure-function investigations of the individual domains or large fragments of DNMT1 and DNMT3A have revealed the molecular basis for their substrate recognition and specificity, intramolecular domain-domain interactions, as well as their crosstalk with other epigenetic mechanisms. These studies highlight a multifaceted regulation for both DNMT1 and DNMT3A/3B, which is essential for the precise establishment and maintenance of lineage-specific DNA methylation patterns in cells. This review summarizes current understanding of the structure and mechanism of DNMT1 and DNMT3A-mediated DNA methylation, with emphasis on the functional cooperation between the methyltransferase and regulatory domains.
Keywords: DNA methyltransferase; DNMT1; DNMT3A; allosteric regulation; autoinhibition; de novo DNA methylation; maintenance DNA methylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures