Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder
- PMID: 30545852
- PMCID: PMC6432922
- DOI: 10.1126/science.aat6576
Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder
Abstract
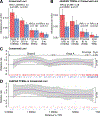
Whole-genome sequencing (WGS) has facilitated the first genome-wide evaluations of the contribution of de novo noncoding mutations to complex disorders. Using WGS, we identified 255,106 de novo mutations among sample genomes from members of 1902 quartet families in which one child, but not a sibling or their parents, was affected by autism spectrum disorder (ASD). In contrast to coding mutations, no noncoding functional annotation category, analyzed in isolation, was significantly associated with ASD. Casting noncoding variation in the context of a de novo risk score across multiple annotation categories, however, did demonstrate association with mutations localized to promoter regions. We found that the strongest driver of this promoter signal emanates from evolutionarily conserved transcription factor binding sites distal to the transcription start site. These data suggest that de novo mutations in promoter regions, characterized by evolutionary and functional signatures, contribute to ASD.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures



References
-
- Heyne HO et al. , De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet 50, 1048–1053 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH110928/MH/NIMH NIH HHS/United States
- R01 MH094714/MH/NIMH NIH HHS/United States
- R21 MH103877/MH/NIMH NIH HHS/United States
- U01 MH103365/MH/NIMH NIH HHS/United States
- R01 MH110905/MH/NIMH NIH HHS/United States
- R01 MH110927/MH/NIMH NIH HHS/United States
- R01 MH109677/MH/NIMH NIH HHS/United States
- U01 MH103346/MH/NIMH NIH HHS/United States
- U01 MH111658/MH/NIMH NIH HHS/United States
- U01 MH103339/MH/NIMH NIH HHS/United States
- R01 MH094400/MH/NIMH NIH HHS/United States
- R21 MH105881/MH/NIMH NIH HHS/United States
- R01 MH110921/MH/NIMH NIH HHS/United States
- R01 MH115957/MH/NIMH NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- U01 MH105575/MH/NIMH NIH HHS/United States
- U01 MH111660/MH/NIMH NIH HHS/United States
- R21 MH109956/MH/NIMH NIH HHS/United States
- U01 MH100239/MH/NIMH NIH HHS/United States
- U01 MH103392/MH/NIMH NIH HHS/United States
- R01 MH111721/MH/NIMH NIH HHS/United States
- U01 MH111661/MH/NIMH NIH HHS/United States
- U01 MH103340/MH/NIMH NIH HHS/United States
- R01 MH109901/MH/NIMH NIH HHS/United States
- R01 HD081256/HD/NICHD NIH HHS/United States
- U01 MH111662/MH/NIMH NIH HHS/United States
- R01 MH110920/MH/NIMH NIH HHS/United States
- R24 HD000836/HD/NICHD NIH HHS/United States
- R56 MH115957/MH/NIMH NIH HHS/United States
- R01 MH109904/MH/NIMH NIH HHS/United States
- R21 MH102791/MH/NIMH NIH HHS/United States
- R01 MH107649/MH/NIMH NIH HHS/United States
- R01 MH105898/MH/NIMH NIH HHS/United States
- R01 MH109900/MH/NIMH NIH HHS/United States
- R21 MH105853/MH/NIMH NIH HHS/United States
- R01 MH109715/MH/NIMH NIH HHS/United States
- R01 MH113557/MH/NIMH NIH HHS/United States
- R01 MH110926/MH/NIMH NIH HHS/United States
- R01 MH049428/MH/NIMH NIH HHS/United States
- P50 MH106934/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

