A Golgi-Released Subpopulation of the Trans-Golgi Network Mediates Protein Secretion in Arabidopsis
- PMID: 30545905
- PMCID: PMC6426420
- DOI: 10.1104/pp.18.01228
A Golgi-Released Subpopulation of the Trans-Golgi Network Mediates Protein Secretion in Arabidopsis
Abstract
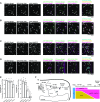
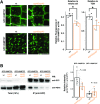
Spatiotemporal coordination of protein trafficking among organelles is essential for eukaryotic cells. The post-Golgi interface, including the trans-Golgi network (TGN), is a pivotal hub for multiple trafficking pathways. The Golgi-released independent TGN (GI-TGN) is a compartment described only in plant cells, and its cellular and physiological roles remain elusive. In Arabidopsis (Arabidopsis thaliana), the SYNTAXIN OF PLANTS (SYP) 4 group Qa-SNARE (soluble N-ethylmaleimide) membrane fusion proteins are shared components of TGN and GI-TGN and regulate secretory and vacuolar transport. Here we reveal that GI-TGNs mediate the transport of the R-SNARE VESICLE-ASSOCIATED MEMBRANE PROTEIN (VAMP) 721 to the plasma membrane. In interactions with a nonadapted powdery mildew pathogen, the SYP4 group of SNAREs is required for the dynamic relocation of VAMP721 to plant-fungus contact sites via GI-TGNs, thereby facilitating complex formation with its cognate SNARE partner PENETRATION1 to restrict pathogen entry. Furthermore, quantitative proteomic analysis of leaf apoplastic fluid revealed constitutive and pathogen-inducible secretion of cell wall-modification enzymes in a SYP4- and VAMP721-dependent manner. Hence, the GI-TGN acts as a transit compartment between the Golgi apparatus and the plasma membrane. We propose a model in which the GA-TGN matures into the GI-TGN and then into secretory vesicles by increasing the abundance of VAMP721-dependent secretory pathway components.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures





Similar articles
-
The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes.Plant J. 2014 Sep;79(5):835-47. doi: 10.1111/tpj.12591. Epub 2014 Jul 23. Plant J. 2014. PMID: 24941879
-
SNARE proteins VAMP721 and VAMP722 mediate the post-Golgi trafficking required for auxin-mediated development in Arabidopsis.Plant J. 2021 Oct;108(2):426-440. doi: 10.1111/tpj.15450. Epub 2021 Aug 31. Plant J. 2021. PMID: 34343378
-
Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells.Cell Struct Funct. 2004 Apr;29(2):49-65. doi: 10.1247/csf.29.49. Cell Struct Funct. 2004. PMID: 15342965
-
RAB GTPases and SNAREs at the trans-Golgi network in plants.J Plant Res. 2022 May;135(3):389-403. doi: 10.1007/s10265-022-01392-x. Epub 2022 Apr 29. J Plant Res. 2022. PMID: 35488138 Free PMC article. Review.
-
Physiological Roles of Plant Post-Golgi Transport Pathways in Membrane Trafficking.Plant Cell Physiol. 2016 Oct;57(10):2013-2019. doi: 10.1093/pcp/pcw149. Epub 2016 Sep 20. Plant Cell Physiol. 2016. PMID: 27649735 Review.
Cited by
-
Duplication of NRAMP3 gene in poplars generated two homologous transporters with distinct functions.Mol Biol Evol. 2022 Jun 14;39(6):msac129. doi: 10.1093/molbev/msac129. Online ahead of print. Mol Biol Evol. 2022. PMID: 35700212 Free PMC article.
-
Differentiation of Trafficking Pathways at Golgi Entry Core Compartments and Post-Golgi Subdomains.Front Plant Sci. 2020 Dec 8;11:609516. doi: 10.3389/fpls.2020.609516. eCollection 2020. Front Plant Sci. 2020. PMID: 33363561 Free PMC article. Review.
-
The sorting of cargo proteins in the plant trans-Golgi network.Front Plant Sci. 2022 Aug 11;13:957995. doi: 10.3389/fpls.2022.957995. eCollection 2022. Front Plant Sci. 2022. PMID: 36035717 Free PMC article. Review.
-
A glossary of plant cell structures: Current insights and future questions.Plant Cell. 2022 Jan 20;34(1):10-52. doi: 10.1093/plcell/koab247. Plant Cell. 2022. PMID: 34633455 Free PMC article. Review.
-
Sweet control: intracellular sorting of glycoproteins in plants.J Exp Bot. 2025 May 27;76(8):2035-2038. doi: 10.1093/jxb/eraf095. J Exp Bot. 2025. PMID: 40425529 Free PMC article.
References
-
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 - PMC - PubMed
-
- Birkenbihl RP, Liu S, Somssich IE (2017) Transcriptional events defining plant immune responses. Curr Opin Plant Biol 38: 1–9 - PubMed
-
- Brito N, Espino JJ, González C (2006) The endo-β-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol Plant Microbe Interact 19: 25–32 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

