Virus-mediated export of chromosomal DNA in plants
- PMID: 30546019
- PMCID: PMC6293997
- DOI: 10.1038/s41467-018-07775-w
Virus-mediated export of chromosomal DNA in plants
Abstract
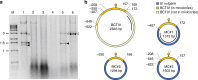
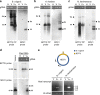
The propensity of viruses to acquire genetic material from relatives and possibly from infected hosts makes them excellent candidates as vectors for horizontal gene transfer. However, virus-mediated acquisition of host genetic material, as deduced from historical events, appears to be rare. Here, we report spontaneous and surprisingly efficient generation of hybrid virus/host DNA molecules in the form of minicircles during infection of Beta vulgaris by Beet curly top Iran virus (BCTIV), a single-stranded DNA virus. The hybrid minicircles replicate, become encapsidated into viral particles, and spread systemically throughout infected plants in parallel with the viral infection. Importantly, when co-infected with BCTIV, B. vulgaris DNA captured in minicircles replicates and is transcribed in other plant species that are sensitive to BCTIV infection. Thus, we have likely documented in real time the initial steps of a possible path of virus-mediated horizontal transfer of chromosomal DNA between plant species.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

