Deciphering bacterial epigenomes using modern sequencing technologies
- PMID: 30546107
- PMCID: PMC6555402
- DOI: 10.1038/s41576-018-0081-3
Deciphering bacterial epigenomes using modern sequencing technologies
Abstract
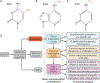
Prokaryotic DNA contains three types of methylation: N6-methyladenine, N4-methylcytosine and 5-methylcytosine. The lack of tools to analyse the frequency and distribution of methylated residues in bacterial genomes has prevented a full understanding of their functions. Now, advances in DNA sequencing technology, including single-molecule, real-time sequencing and nanopore-based sequencing, have provided new opportunities for systematic detection of all three forms of methylated DNA at a genome-wide scale and offer unprecedented opportunities for achieving a more complete understanding of bacterial epigenomes. Indeed, as the number of mapped bacterial methylomes approaches 2,000, increasing evidence supports roles for methylation in regulation of gene expression, virulence and pathogen-host interactions.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

