Comparative genome analysis of marine purple sulfur bacterium Marichromatium gracile YL28 reveals the diverse nitrogen cycle mechanisms and habitat-specific traits
- PMID: 30546119
- PMCID: PMC6292899
- DOI: 10.1038/s41598-018-36160-2
Comparative genome analysis of marine purple sulfur bacterium Marichromatium gracile YL28 reveals the diverse nitrogen cycle mechanisms and habitat-specific traits
Abstract
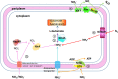
Mangrove ecosystems are characteristic of the high salinity, limited nutrients and S-richness. Marichromatium gracile YL28 (YL28) isolated from mangrove tolerates the high concentrations of nitrite and sulfur compounds and efficiently eliminates them. However, the molecular mechanisms of nitrite and sulfur compounds utilization and the habitat adaptation remain unclear in YL28. We sequenced YL28 genome and further performed the comparative genome analysis in 36 purple bacteria including purple sulfur bacteria (PSB) and purple non-sulfur bacteria (PNSB). YL28 has 6 nitrogen cycle pathways (up to 40 genes), and possibly removes nitrite by denitrification, complete assimilation nitrate reduction and fermentative nitrate reduction (DNRA). Comparative genome analysis showed that more nitrogen utilization genes were detected in PNSB than those in PSB. The partial denitrification pathway and complete assimilation nitrate reduction were reported in PSB and DNRA was reported in purple bacteria for the first time. The three sulfur metabolism genes such as oxidation of sulfide, reversed dissimilatory sulfite reduction and sox system allowed to eliminate toxic sulfur compounds in the mangrove ecosystem. Several unique stress response genes facilitate to the tolerance of the high salinity environment. The CRISPR systems and the transposon components in genomic islands (GIs) likely contribute to the genome plasticity in purple bacteria.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Trüper, H. G. & Pfennig, N. Characterization and identification of the anoxygenic phototrophic bacteria, 1. [Starr, M. P., Stolp, H., Trüper, H. G., Balows, A. & Schlegel, H. G. (eds)] The prokaryotes18, 299–312. (Springer, Berlin, Heidelberg, 1981).
-
- Zhao JY, et al. Identification and characterization of a purple sulfur bacterium from mangrove with rhodopin as predominant carotenoid. Acta Microbiologica Sinica. 2011;51(10):1318–1325. - PubMed
-
- Jiang P, Hong X, Zhao CG, Yang SP. Reciprocal transformation of Inorganic nitrogen by Resting Cells of Marichromatium gracile YL28. Journal of Huaqiao University. 2015;36(2):45–52.
-
- Jiang P, Zhao CG, Jia YQ, Yang SP. Inorganic nitrogen removal by a marine purple sulfur bacterium capable of growth on nitrite as sole nitrogen source. Microbiology China. 2014;41(5):824–831.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

