Molecular Mechanisms of Bacterial Bioluminescence
- PMID: 30546856
- PMCID: PMC6279958
- DOI: 10.1016/j.csbj.2018.11.003
Molecular Mechanisms of Bacterial Bioluminescence
Abstract
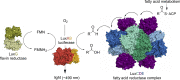
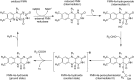
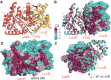
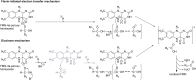
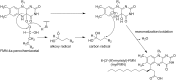
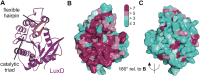
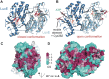
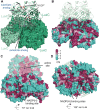
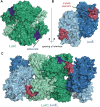
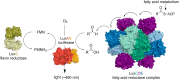
Bioluminescence refers to the production of light by living organisms. Bioluminescent bacteria with a variety of bioluminescence emission characteristics have been identified in Vibrionaceae, Shewanellaceae and Enterobacteriaceae. Bioluminescent bacteria are mainly found in marine habitats and they are either free-floating, sessile or have specialized to live in symbiosis with other marine organisms. On the molecular level, bioluminescence is enabled by a cascade of chemical reactions catalyzed by enzymes encoded by the lux operon with the gene order luxCDABEG. The luxA and luxB genes encode the α- and β- subunits, respectively, of the enzyme luciferase producing the light emitting species. LuxC, luxD and luxE constitute the fatty acid reductase complex, responsible for the synthesis of the long-chain aldehyde substrate and luxG encodes a flavin reductase. In bacteria, the heterodimeric luciferase catalyzes the monooxygenation of long-chain aliphatic aldehydes to the corresponding acids utilizing reduced FMN and molecular oxygen. The energy released as a photon results from an excited state flavin-4a-hydroxide, emitting light centered around 490 nm. Advances in the mechanistic understanding of bacterial bioluminescence have been spurred by the structural characterization of protein encoded by the lux operon. However, the number of available crystal structures is limited to LuxAB (Vibrio harveyi), LuxD (Vibrio harveyi) and LuxF (Photobacterium leiognathi). Based on the crystal structure of LuxD and homology models of LuxC and LuxE, we provide a hypothetical model of the overall structure of the LuxCDE fatty acid reductase complex that is in line with biochemical observations.
Keywords: Bacterial bioluminescence; FMN; Fatty acid reductase complex; Luciferase; Luciferin; Structure-function relationships; lux operon.
Figures

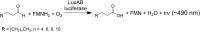
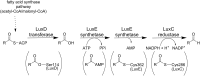
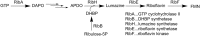


Similar articles
-
Biochemistry and genetics of bacterial bioluminescence.Adv Biochem Eng Biotechnol. 2014;144:37-64. doi: 10.1007/978-3-662-43385-0_2. Adv Biochem Eng Biotechnol. 2014. PMID: 25084994 Review.
-
Characteristic analysis of the luxG gene encoding the probable flavin reductase that resides in the lux operon of Photobacterium leiognathi.Biochem Biophys Res Commun. 1998 May 19;246(2):446-52. doi: 10.1006/bbrc.1998.8641. Biochem Biophys Res Commun. 1998. PMID: 9610381
-
Nucleotide sequence and functional analysis of the luxE gene encoding acyl-protein synthetase of the lux operon from Photobacterium leiognathi.Biochem Biophys Res Commun. 1996 Nov 21;228(3):764-73. doi: 10.1006/bbrc.1996.1729. Biochem Biophys Res Commun. 1996. PMID: 8941351
-
The impact of LuxF on light intensity in bacterial bioluminescence.J Photochem Photobiol B. 2020 Jun;207:111881. doi: 10.1016/j.jphotobiol.2020.111881. Epub 2020 Apr 9. J Photochem Photobiol B. 2020. PMID: 32325406
-
Mechanisms and applications of bacterial luciferase and its auxiliary enzymes.Arch Biochem Biophys. 2025 Mar;765:110307. doi: 10.1016/j.abb.2025.110307. Epub 2025 Jan 15. Arch Biochem Biophys. 2025. PMID: 39824239 Review.
Cited by
-
From macro to micro: a combined bioluminescence-fluorescence approach to monitor bacterial localization.Environ Microbiol. 2021 Apr;23(4):2070-2085. doi: 10.1111/1462-2920.15296. Epub 2021 Jan 22. Environ Microbiol. 2021. PMID: 33103833 Free PMC article.
-
Blue light promotes zero-valent sulfur production in a deep-sea bacterium.EMBO J. 2023 Jun 15;42(12):e112514. doi: 10.15252/embj.2022112514. Epub 2023 Mar 22. EMBO J. 2023. PMID: 36946144 Free PMC article.
-
Bacterial Lighthouses-Real-Time Detection of Yersinia enterocolitica by Quorum Sensing.Biosensors (Basel). 2021 Dec 16;11(12):517. doi: 10.3390/bios11120517. Biosensors (Basel). 2021. PMID: 34940274 Free PMC article.
-
The Assessment on Synergistic Activity of Ebselen and Silver Ion Against Yersinia pseudotuberculosis.Front Microbiol. 2022 Jul 25;13:963901. doi: 10.3389/fmicb.2022.963901. eCollection 2022. Front Microbiol. 2022. PMID: 35958130 Free PMC article.
-
Generation of Fluorescent Bacteria with the Genes Coding for Lumazine Protein and Riboflavin Biosynthesis.Sensors (Basel). 2021 Jun 30;21(13):4506. doi: 10.3390/s21134506. Sensors (Basel). 2021. PMID: 34209387 Free PMC article.
References
-
- Widder E.A. Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science. 2010;328:704–708. 80- - PubMed
-
- Dunlap P. In: Bioluminescence, microbial. Schaechter M., editor. Elsevier; Oxford: 2009. pp. 45–61. Encycl Microbiol.
-
- Haddock S.H.D., Moline M.A., Case J.F. Bioluminescence in the sea. Ann Rev Mar Sci. 2010;2:443–493. - PubMed
-
- Hastings J.W., Potrikus C.J., Gupta S.C., Kurfürst M., Makemson J.C. Biochemistry and physiology of bioluminescent bacteria. Adv Microb Physiol. 1985;26:235–291. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
