Biochemical Characterization of Aspergillus fumigatus AroH, a Putative Aromatic Amino Acid Aminotransferase
- PMID: 30547035
- PMCID: PMC6279937
- DOI: 10.3389/fmolb.2018.00104
Biochemical Characterization of Aspergillus fumigatus AroH, a Putative Aromatic Amino Acid Aminotransferase
Abstract
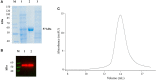
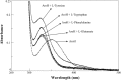
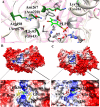
The rise in the frequency of nosocomial infections is becoming a major problem for public health, in particular in immunocompromised patients. Aspergillus fumigatus is an opportunistic fungus normally present in the environment directly responsible for lethal invasive infections. Recent results suggest that the metabolic pathways related to amino acid metabolism can regulate the fungus-host interaction and that an important role is played by enzymes involved in the catabolism of L-tryptophan. In particular, in A. fumigatus L-tryptophan regulates Aro genes. Among them, AroH encodes a putative pyridoxal 5'-phosphate-dependent aminotransferase. Here we analyzed the biochemical features of recombinant purified AroH by spectroscopic and kinetic analyses corroborated by in silico studies. We found that the protein is dimeric and tightly binds the coenzyme forming a deprotonated internal aldimine in equilibrium with a protonated ketoenamine form. By setting up a new rapid assay method, we measured the kinetic parameters for the overall transamination of substrates and we demonstrated that AroH behaves as an aromatic amino acid aminotransferase, but also accepts L-kynurenine and α-aminoadipate as amino donors. Interestingly, computational approaches showed that the predicted overall fold and active site topology of the protein are similar to those of its yeast ortholog, albeit with some differences in the regions at the entrance of the active site, which could possibly influence substrate specificity. Should targeting fungal metabolic adaptation be of therapeutic value, the results of the present study may pave the way to the design of specific AroH modulators as potential novel agents at the host/fungus interface.
Keywords: aromatic amino acid aminotransferase; enzymatic assay; enzyme kinetics; enzyme spectroscopy; fungal infection; homology modeling; pyridoxal 5'-phosphate.
Figures





Similar articles
-
Crystal structure of Aspergillus fumigatus AroH, an aromatic amino acid aminotransferase.Proteins. 2022 Feb;90(2):435-442. doi: 10.1002/prot.26234. Epub 2021 Sep 16. Proteins. 2022. PMID: 34495558 Free PMC article.
-
Biochemical characterization of plant aromatic aminotransferases.Methods Enzymol. 2023;680:35-83. doi: 10.1016/bs.mie.2022.07.034. Epub 2022 Sep 7. Methods Enzymol. 2023. PMID: 36710018
-
Escherichia coli aromatic amino acid aminotransferase: characterization and comparison with aspartate aminotransferase.Biochemistry. 1993 Nov 16;32(45):12229-39. doi: 10.1021/bi00096a036. Biochemistry. 1993. PMID: 8218300
-
Structure and mechanism of kynureninase.Arch Biochem Biophys. 2014 Feb 15;544:69-74. doi: 10.1016/j.abb.2013.10.020. Epub 2013 Nov 4. Arch Biochem Biophys. 2014. PMID: 24200862 Review.
-
The role of glutamine transaminase K (GTK) in sulfur and alpha-keto acid metabolism in the brain, and in the possible bioactivation of neurotoxicants.Neurochem Int. 2004 Jun;44(8):557-77. doi: 10.1016/j.neuint.2003.12.002. Neurochem Int. 2004. PMID: 15016471 Review.
Cited by
-
Tryptophan Co-Metabolism at the Host-Pathogen Interface.Front Immunol. 2020 Jan 29;11:67. doi: 10.3389/fimmu.2020.00067. eCollection 2020. Front Immunol. 2020. PMID: 32082324 Free PMC article. No abstract available.
-
Aspergillus fumigatus tryptophan metabolic route differently affects host immunity.Cell Rep. 2021 Jan 26;34(4):108673. doi: 10.1016/j.celrep.2020.108673. Cell Rep. 2021. PMID: 33503414 Free PMC article.
-
Optimized psilocybin production in tryptophan catabolism-repressed fungi.Microb Biotechnol. 2024 Nov;17(11):e70039. doi: 10.1111/1751-7915.70039. Microb Biotechnol. 2024. PMID: 39487767 Free PMC article.
-
Modeling Approaches Reveal New Regulatory Networks in Aspergillus fumigatus Metabolism.J Fungi (Basel). 2020 Jul 14;6(3):108. doi: 10.3390/jof6030108. J Fungi (Basel). 2020. PMID: 32674323 Free PMC article.
-
Crystal structure of Aspergillus fumigatus AroH, an aromatic amino acid aminotransferase.Proteins. 2022 Feb;90(2):435-442. doi: 10.1002/prot.26234. Epub 2021 Sep 16. Proteins. 2022. PMID: 34495558 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

