Human D-Amino Acid Oxidase: Structure, Function, and Regulation
- PMID: 30547037
- PMCID: PMC6279847
- DOI: 10.3389/fmolb.2018.00107
Human D-Amino Acid Oxidase: Structure, Function, and Regulation
Abstract
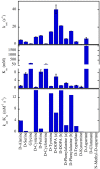
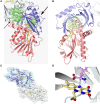
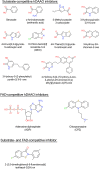
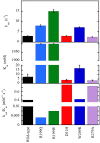
D-Amino acid oxidase (DAAO) is an FAD-containing flavoenzyme that catalyzes with absolute stereoselectivity the oxidative deamination of all natural D-amino acids, the only exception being the acidic ones. This flavoenzyme plays different roles during evolution and in different tissues in humans. Its three-dimensional structure is well conserved during evolution: minute changes are responsible for the functional differences between enzymes from microorganism sources and those from humans. In recent years several investigations focused on human DAAO, mainly because of its role in degrading the neuromodulator D-serine in the central nervous system. D-Serine is the main coagonist of N-methyl D-aspartate receptors, i.e., excitatory amino acid receptors critically involved in main brain functions and pathologic conditions. Human DAAO possesses a weak interaction with the FAD cofactor; thus, in vivo it should be largely present in the inactive, apoprotein form. Binding of active-site ligands and the substrate stabilizes flavin binding, thus pushing the acquisition of catalytic competence. Interestingly, the kinetic efficiency of the enzyme on D-serine is very low. Human DAAO interacts with various proteins, in this way modulating its activity, targeting, and cell stability. The known properties of human DAAO suggest that its activity must be finely tuned to fulfill a main physiological function such as the control of D-serine levels in the brain. At present, studies are focusing on the epigenetic modulation of human DAAO expression and the role of post-translational modifications on its main biochemical properties at the cellular level.
Keywords: D-amino acid oxidase; D-serine; NMDA receptor; structure-function relationships; substrate specificity.
Figures

Similar articles
-
Biochemical Properties of Human D-Amino Acid Oxidase.Front Mol Biosci. 2017 Dec 15;4:88. doi: 10.3389/fmolb.2017.00088. eCollection 2017. Front Mol Biosci. 2017. PMID: 29326945 Free PMC article.
-
Is rat an appropriate animal model to study the involvement of D-serine catabolism in schizophrenia? Insights from characterization of D-amino acid oxidase.FEBS J. 2011 Nov;278(22):4362-73. doi: 10.1111/j.1742-4658.2011.08354.x. Epub 2011 Oct 10. FEBS J. 2011. PMID: 21981077
-
Targeting D-Amino Acid Oxidase (DAAO) for the Treatment of Schizophrenia: Rationale and Current Status of Research.CNS Drugs. 2022 Nov;36(11):1143-1153. doi: 10.1007/s40263-022-00959-5. Epub 2022 Oct 4. CNS Drugs. 2022. PMID: 36194364
-
D-amino acid oxidase inhibitors as a novel class of drugs for schizophrenia therapy.Curr Pharm Des. 2013;19(14):2499-511. doi: 10.2174/1381612811319140002. Curr Pharm Des. 2013. PMID: 23116391 Review.
-
Biochemical Properties of Human D-amino Acid Oxidase Variants and Their Potential Significance in Pathologies.Front Mol Biosci. 2018 Jun 12;5:55. doi: 10.3389/fmolb.2018.00055. eCollection 2018. Front Mol Biosci. 2018. PMID: 29946548 Free PMC article. Review.
Cited by
-
Diagnosing Alzheimer's Disease Specifically and Sensitively With pLG72 and Cystine/Glutamate Antiporter SLC7A11 AS Blood Biomarkers.Int J Neuropsychopharmacol. 2023 Jan 19;26(1):1-8. doi: 10.1093/ijnp/pyac053. Int J Neuropsychopharmacol. 2023. PMID: 35986919 Free PMC article.
-
Structural Determinants of the Specific Activities of an L-Amino Acid Oxidase from Pseudoalteromonas luteoviolacea CPMOR-1 with Broad Substrate Specificity.Molecules. 2022 Jul 24;27(15):4726. doi: 10.3390/molecules27154726. Molecules. 2022. PMID: 35897902 Free PMC article.
-
The D-amino acid oxidase-carbon nanotubes: evaluation of cytotoxicity and biocompatibility of a potential anticancer nanosystem.3 Biotech. 2023 Jul;13(7):243. doi: 10.1007/s13205-023-03568-1. Epub 2023 Jun 19. 3 Biotech. 2023. PMID: 37346390 Free PMC article.
-
Dissecting in vivo and in vitro redox responses using chemogenetics.Free Radic Biol Med. 2021 Dec;177:360-369. doi: 10.1016/j.freeradbiomed.2021.11.006. Epub 2021 Nov 6. Free Radic Biol Med. 2021. PMID: 34752919 Free PMC article. Review.
-
Mammalian D-Cysteine controls insulin secretion in the pancreas.Mol Metab. 2024 Dec;90:102043. doi: 10.1016/j.molmet.2024.102043. Epub 2024 Oct 3. Mol Metab. 2024. PMID: 39368613 Free PMC article.
References
-
- Adage T., Trillat A. C., Quattropani A., Perrin D., Cavarec L., Shaw J., et al. . (2008). In vitro and in vivo pharmacological profile of AS057278, a selective D-amino acid oxidase inhibitor with potential anti-psychotic properties. Eur. Neuropsychopharmacol. 18, 200–214. 10.1016/j.euroneuro.2007.06.006 - DOI - PubMed
-
- Arroyo M., Menéndez M., García J. L., Campillo N., Hormigo D., de la Mata I., et al. . (2007). The role of cofactor binding in tryptophan accessibility and conformational stability of His-tagged D-amino acid oxidase from Trigonopsis variabilis. Biochim. Biophys. Acta 1774, 556–565. 10.1016/j.bbapap.2007.03.009 - DOI - PubMed
-
- Beato C., Pecchini C., Cocconcelli C., Campanini B., Marchetti M., Pieroni M., et al. . (2016). Cyclopropane derivatives as potential human serine racemase inhibitors: unveiling novel insights into a difficult target. J. Enzyme. Inhib. Med. Chem. 31, 645–652. 10.3109/14756366.2015.1057720 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

