Site-specific 2D IR spectroscopy: a general approach for the characterization of protein dynamics with high spatial and temporal resolution
- PMID: 30548035
- PMCID: PMC6360950
- DOI: 10.1039/c8cp06146g
Site-specific 2D IR spectroscopy: a general approach for the characterization of protein dynamics with high spatial and temporal resolution
Abstract
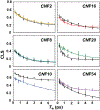
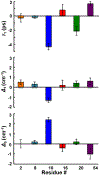
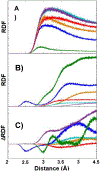
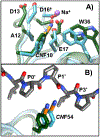
The conformational heterogeneity and dynamics of protein side chains contribute to function, but investigating exactly how is hindered by experimental challenges arising from the fast timescales involved and the spatial heterogeneity of protein structures. The potential of two-dimensional infrared (2D IR) spectroscopy for measuring conformational heterogeneity and dynamics with unprecedented spatial and temporal resolution has motivated extensive effort to develop amino acids with functional groups that have frequency-resolved absorptions to serve as probes of their protein microenvironments. We demonstrate the full advantage of the approach by selective incorporation of the probe p-cyanophenylalanine at six distinct sites in a Src homology 3 domain and the application of 2D IR spectroscopy to site-specifically characterize heterogeneity and dynamics and their contribution to cognate ligand binding. The approach revealed a wide range of microenvironments and distinct responses to ligand binding, including at the three adjacent, conserved aromatic residues that form the recognition surface of the protein. Molecular dynamics simulations performed for all the labeled proteins provide insight into the underlying heterogeneity and dynamics. Similar application of 2D IR spectroscopy and site-selective probe incorporation will allow for the characterization of heterogeneity and dynamics of other proteins, how heterogeneity and dynamics are affected by solvation and local structure, and how they might contribute to biological function.
Conflict of interest statement
Conflicts of interest
The authors declare no conflict of interest.
Figures


Similar articles
-
Transparent window 2D IR spectroscopy of proteins.J Chem Phys. 2021 Jul 28;155(4):040903. doi: 10.1063/5.0052628. J Chem Phys. 2021. PMID: 34340394 Free PMC article. Review.
-
Site-Specific 1D and 2D IR Spectroscopy to Characterize the Conformations and Dynamics of Protein Molecular Recognition.J Phys Chem B. 2019 May 2;123(17):3551-3566. doi: 10.1021/acs.jpcb.9b00969. Epub 2019 Mar 21. J Phys Chem B. 2019. PMID: 30848912 Free PMC article.
-
Resolution of Site-Specific Conformational Heterogeneity in Proline-Rich Molecular Recognition by Src Homology 3 Domains.J Am Chem Soc. 2016 Feb 3;138(4):1130-3. doi: 10.1021/jacs.5b11999. Epub 2016 Jan 25. J Am Chem Soc. 2016. PMID: 26784847 Free PMC article.
-
Site-selective Characterization of Src Homology 3 Domain Molecular Recognition with Cyanophenylalanine Infrared Probes.Anal Methods. 2015;7:7234-7241. doi: 10.1039/C5AY00523J. Epub 2015 Apr 8. Anal Methods. 2015. PMID: 26491469 Free PMC article.
-
Ultrafast structural molecular dynamics investigated with 2D infrared spectroscopy methods.Top Curr Chem (Cham). 2017 Oct 25;375(6):86. doi: 10.1007/s41061-017-0172-1. Top Curr Chem (Cham). 2017. PMID: 29071445 Review.
Cited by
-
Facile Generation of Cyanoselenocysteine as a Vibrational Label for Measuring Protein Dynamics on Longer Time Scales by 2D IR Spectroscopy.Anal Chem. 2025 Jan 28;97(3):1673-1680. doi: 10.1021/acs.analchem.4c04689. Epub 2025 Jan 10. Anal Chem. 2025. PMID: 39791917
-
Transparent window 2D IR spectroscopy of proteins.J Chem Phys. 2021 Jul 28;155(4):040903. doi: 10.1063/5.0052628. J Chem Phys. 2021. PMID: 34340394 Free PMC article. Review.
-
Protein Dynamics by Two-Dimensional Infrared Spectroscopy.Annu Rev Anal Chem (Palo Alto Calif). 2021 Jul 27;14(1):299-321. doi: 10.1146/annurev-anchem-091520-091009. Annu Rev Anal Chem (Palo Alto Calif). 2021. PMID: 34314221 Free PMC article.
-
Site-Specific 1D and 2D IR Spectroscopy to Characterize the Conformations and Dynamics of Protein Molecular Recognition.J Phys Chem B. 2019 May 2;123(17):3551-3566. doi: 10.1021/acs.jpcb.9b00969. Epub 2019 Mar 21. J Phys Chem B. 2019. PMID: 30848912 Free PMC article.
-
Anisotropic dynamics of an interfacial enzyme active site observed using tethered substrate analogs and ultrafast 2D IR spectroscopy.J Chem Phys. 2023 Oct 28;159(16):165101. doi: 10.1063/5.0167991. J Chem Phys. 2023. PMID: 37870142 Free PMC article.
References
-
- Palmer AG III, Annu. Rev. Biophys. Biomol. Struct, 2001, 30, 129–155. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

