Impact of differential DNA methylation on transgene expression in cotton (Gossypium hirsutum L.) events generated by targeted sequence insertion
- PMID: 30549163
- PMCID: PMC6576080
- DOI: 10.1111/pbi.13049
Impact of differential DNA methylation on transgene expression in cotton (Gossypium hirsutum L.) events generated by targeted sequence insertion
Abstract
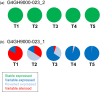
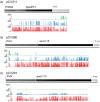
Targeted Genome Optimization (TGO) using site-specific nucleases to introduce a DNA double-strand break (DSB) at a specific target locus has broadened the options available to breeders for generation and combination of multiple traits. The use of targeted DNA cleavage in combination with homologous recombination (HR)-mediated repair, enabled the precise targeted insertion of additional trait genes (2mepsps, hppd, axmi115) at a pre-existing transgenic locus in cotton. Here we describe the expression and epigenome analyses of cotton Targeted Sequence Insertion (TSI) events over generations. In a subset of events, we observed variability in the level of transgene (hppd, axmi115) expression between independent but genetically identical TSI events. Transgene expression could also be differential within single events and variable over generations. This expression variability and silencing occurred independently of the transgene sequence and could be attributed to DNA methylation that was further linked to different DNA methylation mechanisms. The trigger(s) of transgene DNA methylation remains elusive but we hypothesize that targeted DSB induction and repair could be a potential trigger for DNA methylation.
Keywords: DNA methylation; DNA methylation mechanisms; cotton; differential silencing; epigenome analyses; expression variability; gene targeting; targeted sequence insertion; transgene expression; transgene silencing.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures





References
-
- Ainley, W.M. , Sastry‐Dent, L. , Welter, M.E. , Murray, M.G. , Zeitler, B. , Amora, R. , Corbin, D.R. et al. (2013) Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

