Guided cobamide biosynthesis for heterologous production of reductive dehalogenases
- PMID: 30549216
- PMCID: PMC6389850
- DOI: 10.1111/1751-7915.13339
Guided cobamide biosynthesis for heterologous production of reductive dehalogenases
Abstract
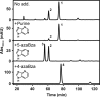
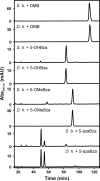
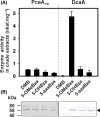
Cobamides (Cbas) are essential cofactors of reductive dehalogenases (RDases) in organohalide-respiring bacteria (OHRB). Changes in the Cba structure can influence RDase function. Here, we report on the cofactor versatility or selectivity of Desulfitobacterium RDases produced either in the native organism or heterologously. The susceptibility of Desulfitobacterium hafniense strain DCB-2 to guided Cba biosynthesis (i.e. incorporation of exogenous Cba lower ligand base precursors) was analysed. Exogenous benzimidazoles, azabenzimidazoles and 4,5-dimethylimidazole were incorporated by the organism into Cbas. When the type of Cba changed, no effect on the turnover rate of the 3-chloro-4-hydroxy-phenylacetate-converting enzyme RdhA6 and the 3,5-dichlorophenol-dehalogenating enzyme RdhA3 was observed. The impact of the amendment of Cba lower ligand precursors on RDase function was also investigated in Shimwellia blattae, the Cba producer used for the heterologous production of Desulfitobacterium RDases. The recombinant tetrachloroethene RDase (PceAY51 ) appeared to be non-selective towards different Cbas. However, the functional production of the 1,2-dichloroethane-dihaloeliminating enzyme (DcaA) of Desulfitobacterium dichloroeliminans was completely prevented in cells producing 5,6-dimethylbenzimidazolyl-Cba, but substantially enhanced in cells that incorporated 5-methoxybenzimidazole into the Cba cofactor. The results of the study indicate the utilization of a range of different Cbas by Desulfitobacterium RDases with selected representatives apparently preferring distinct Cbas.
© 2018 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Understanding the sources, function, and irreplaceable role of cobamides in organohalide-respiring bacteria.Front Microbiol. 2024 Jul 30;15:1435674. doi: 10.3389/fmicb.2024.1435674. eCollection 2024. Front Microbiol. 2024. PMID: 39139376 Free PMC article. Review.
-
Subtle changes in the active site architecture untangled overlapping substrate ranges and mechanistic differences of two reductive dehalogenases.FEBS J. 2017 Oct;284(20):3520-3535. doi: 10.1111/febs.14258. Epub 2017 Sep 15. FEBS J. 2017. PMID: 28869789
-
Functional heterologous production of reductive dehalogenases from Desulfitobacterium hafniense strains.Appl Environ Microbiol. 2014 Jul;80(14):4313-22. doi: 10.1128/AEM.00881-14. Epub 2014 May 9. Appl Environ Microbiol. 2014. PMID: 24814779 Free PMC article.
-
Purinyl-cobamide is a native prosthetic group of reductive dehalogenases.Nat Chem Biol. 2018 Jan;14(1):8-14. doi: 10.1038/nchembio.2512. Epub 2017 Nov 6. Nat Chem Biol. 2018. PMID: 29106396 Free PMC article.
-
The organohalide-respiring bacterium Sulfurospirillum multivorans: a natural source for unusual cobamides.World J Microbiol Biotechnol. 2017 May;33(5):93. doi: 10.1007/s11274-017-2258-x. Epub 2017 Apr 10. World J Microbiol Biotechnol. 2017. PMID: 28397170 Review.
Cited by
-
Burning question: Rethinking organohalide degradation strategy for bioremediation applications.Microb Biotechnol. 2024 Aug;17(8):e14539. doi: 10.1111/1751-7915.14539. Microb Biotechnol. 2024. PMID: 39075849 Free PMC article. Review.
-
Inter-bacterial mutualism promoted by public goods in a system characterized by deterministic temperature variation.Nat Commun. 2023 Sep 5;14(1):5394. doi: 10.1038/s41467-023-41224-7. Nat Commun. 2023. PMID: 37669961 Free PMC article.
-
Understanding the sources, function, and irreplaceable role of cobamides in organohalide-respiring bacteria.Front Microbiol. 2024 Jul 30;15:1435674. doi: 10.3389/fmicb.2024.1435674. eCollection 2024. Front Microbiol. 2024. PMID: 39139376 Free PMC article. Review.
-
Cofactor Selectivity in Methylmalonyl Coenzyme A Mutase, a Model Cobamide-Dependent Enzyme.mBio. 2019 Sep 24;10(5):e01303-19. doi: 10.1128/mBio.01303-19. mBio. 2019. PMID: 31551329 Free PMC article.
-
Stoichiometry of the Gene Products From the Tetrachloroethene Reductive Dehalogenase Operon pceABCT.Front Microbiol. 2022 Feb 23;13:838026. doi: 10.3389/fmicb.2022.838026. eCollection 2022. Front Microbiol. 2022. PMID: 35283847 Free PMC article.
References
-
- Andres, S. , Wiezer, A. , Bendfeldt, H. , Waschkowitz, T. , Toeche‐Mittler, C. , and Daniel, R. (2004) Insights into the genome of the enteric bacterium Escherichia blattae: cobalamin (B12) biosynthesis, B12‐dependent reactions, and inactivation of the gene region encoding B12‐dependent glycerol dehydratase by a new mu‐like prophage. J Mol Microbiol Biotechnol 8: 150–168. - PubMed
-
- Banerjee, R. , and Ragsdale, S.W. (2003) The many faces of vitamin B12: catalysis by cobalamin‐dependent enzymes. Annu Rev Biochem 72: 209–247. - PubMed
-
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72: 248–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

