xCas9 expands the scope of genome editing with reduced efficiency in rice
- PMID: 30549238
- PMCID: PMC6419569
- DOI: 10.1111/pbi.13053
xCas9 expands the scope of genome editing with reduced efficiency in rice
Keywords: genome editing; rice; xCas9.
Figures
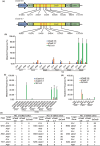
References
-
- Hu, X. , Wang, C. , Fu, Y. , Liu, Q. , Jiao, X. and Wang, K. (2016) Expanding the range of CRISPR/Cas9 genome editing in rice. Mol Plant, 9, 943–945. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

