RAD-ical New Insights into RAD51 Regulation
- PMID: 30551670
- PMCID: PMC6316741
- DOI: 10.3390/genes9120629
RAD-ical New Insights into RAD51 Regulation
Abstract
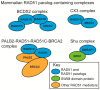
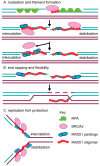
The accurate repair of DNA is critical for genome stability and cancer prevention. DNA double-strand breaks are one of the most toxic lesions; however, they can be repaired using homologous recombination. Homologous recombination is a high-fidelity DNA repair pathway that uses a homologous template for repair. One central HR step is RAD51 nucleoprotein filament formation on the single-stranded DNA ends, which is a step required for the homology search and strand invasion steps of HR. RAD51 filament formation is tightly controlled by many positive and negative regulators, which are collectively termed the RAD51 mediators. The RAD51 mediators function to nucleate, elongate, stabilize, and disassemble RAD51 during repair. In model organisms, RAD51 paralogs are RAD51 mediator proteins that structurally resemble RAD51 and promote its HR activity. New functions for the RAD51 paralogs during replication and in RAD51 filament flexibility have recently been uncovered. Mutations in the human RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3, and SWSAP1) are found in a subset of breast and ovarian cancers. Despite their discovery three decades ago, few advances have been made in understanding the function of the human RAD51 paralogs. Here, we discuss the current perspective on the in vivo and in vitro function of the RAD51 paralogs, and their relationship with cancer in vertebrate models.
Keywords: BRCA1; BRCA2; RAD51 paralogs; RAD51B; RAD51C; RAD51D; Shu complex; XRCC2; XRCC3; cancer; homologous recombination.
Conflict of interest statement
The authors declare no conflicts of interest
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

