Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons
- PMID: 30551672
- PMCID: PMC6316676
- DOI: 10.3390/genes9120630
Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons
Abstract
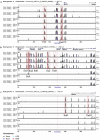
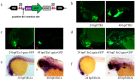
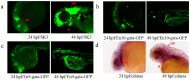
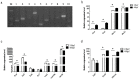
Although transposon-mediated enhancer trapping (ET) is successfully applied in diverse models, the efficiency of various transposon systems varies significantly, and little information is available regarding efficiency of enhancer trapping by various transposons in zebrafish. Most potential enhancers (Ens) still lack evidence of actual En activity. Here, we compared the differences in ET efficiency between sleeping beauty (SB), piggyBac (PB) and Tol2 transposons. Tol2 represented the highest germline transfer efficiencies at 55.56% (NF0 = 165), followed by SB (38.36%, NF0 = 151) and PB (32.65%, NF0 = 149). ET lines generated by the Tol2 transposon tended to produce offspring with a single expression pattern per line, while PB and SB tended to generate embryos with multiple expression patterns. In our tests, 10 putative Ens (En1⁻10) were identified by splinkerette PCR and comparative genomic analysis. Combining the GFP expression profiles and mRNA expression patterns revealed that En1 and En2 may be involved in regulation of the expression of dlx1a and dlx2a, while En6 may be involved in regulation of the expression of line TK4 transgene and rps26, and En7 may be involved in the regulation of the expression of wnt1 and wnt10b. Most identified Ens were found to be transcribed in zebrafish embryos, and their regulatory function may involve eRNAs.
Keywords: Sleeping Beauty; Tol2; enhancer; enhancer trapping; piggyBac; zebrafish.
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures



References
-
- Soldner F., Stelzer Y., Shivalila C.S., Abraham B.J., Latourelle J.C., Barrasa M.I., Goldmann J., Myers R.H., Young R.A., Jaenisch R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95. doi: 10.1038/nature17939. - DOI - PMC - PubMed
Grants and funding
- None/the Postgraduate Research & Practical Innovation Program of Jiangsu Province
- 2018ZX0801003B/Major projects of national genetically modified organism breeding
- None/the Priority Academic Program Development of Jiangsu Higher Education Institutions
- 31671313/the Natural Science Foundation of China
- Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

