Inference of the HIV-1 VRC01 Antibody Lineage Unmutated Common Ancestor Reveals Alternative Pathways to Overcome a Key Glycan Barrier
- PMID: 30552024
- PMCID: PMC6303191
- DOI: 10.1016/j.immuni.2018.10.015
Inference of the HIV-1 VRC01 Antibody Lineage Unmutated Common Ancestor Reveals Alternative Pathways to Overcome a Key Glycan Barrier
Abstract
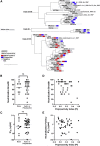
Elicitation of VRC01-class broadly neutralizing antibodies (bnAbs) is an appealing approach for a preventative HIV-1 vaccine. Despite extensive investigations, strategies to induce VRC01-class bnAbs and overcome the barrier posed by the envelope N276 glycan have not been successful. Here, we inferred a high-probability unmutated common ancestor (UCA) of the VRC01 lineage and reconstructed the stages of lineage maturation. Env immunogens designed on reverted VRC01-class bnAbs bound to VRC01 UCA with affinity sufficient to activate naive B cells. Early mutations defined maturation pathways toward limited or broad neutralization, suggesting that focusing the immune response is likely required to steer B cell maturation toward the development of neutralization breadth. Finally, VRC01 lineage bnAbs with long CDR H3s overcame the HIV-1 N276 glycan barrier without shortening their CDR L1, revealing a solution for broad neutralization in which the heavy chain, not CDR L1, is the determinant to accommodate the N276 glycan.
Keywords: B lymphocytes; HIV-1; cell lineage; clonal evolution; glycans; immunogen design; indel mutation; monoclonal antibody VRC01; neutralizing antibodies; vaccines.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Benjamin D., Magrath I.T., Maguire R., Janus C., Todd H.D., Parsons R.G. Immunoglobulin secretion by cell lines derived from African and American undifferentiated lymphomas of Burkitt’s and non-Burkitt’s type. J. Immunol. 1982;129:1336–1342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

