Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study
- PMID: 30552067
- PMCID: PMC6314966
- DOI: 10.1016/S2213-2600(18)30389-8
Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study
Abstract
Background: Few genetic studies that focus on moderate-to-severe asthma exist. We aimed to identity novel genetic variants associated with moderate-to-severe asthma, see whether previously identified genetic variants for all types of asthma contribute to moderate-to-severe asthma, and provide novel mechanistic insights using expression analyses in patients with asthma.
Methods: In this genome-wide association study, we used a two-stage case-control design. In stage 1, we genotyped patient-level data from two UK cohorts (the Genetics of Asthma Severity and Phenotypes [GASP] initiative and the Unbiased BIOmarkers in PREDiction of respiratory disease outcomes [U-BIOPRED] project) and used data from the UK Biobank to collect patient-level genomic data for cases and controls of European ancestry in a 1:5 ratio. Cases were defined as having moderate-to-severe asthma if they were taking appropriate medication or had been diagnosed by a doctor. Controls were defined as not having asthma, rhinitis, eczema, allergy, emphysema, or chronic bronchitis as diagnosed by a doctor. For stage 2, an independent cohort of cases and controls (1:5) was selected from the UK Biobank only, with no overlap with stage 1 samples. In stage 1 we undertook a genome-wide association study of moderate-to-severe asthma, and in stage 2 we followed up independent variants that reached the significance threshold of p less than 1 × 10-6 in stage 1. We set genome-wide significance at p less than 5 × 10-8. For novel signals, we investigated their effect on all types of asthma (mild, moderate, and severe). For all signals meeting genome-wide significance, we investigated their effect on gene expression in patients with asthma and controls.
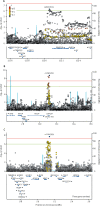
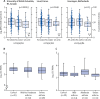
Findings: We included 5135 cases and 25 675 controls for stage 1, and 5414 cases and 21 471 controls for stage 2. We identified 24 genome-wide significant signals of association with moderate-to-severe asthma, including several signals in innate or adaptive immune-response genes. Three novel signals were identified: rs10905284 in GATA3 (coded allele A, odds ratio [OR] 0·90, 95% CI 0·88-0·93; p=1·76 × 10-10), rs11603634 in the MUC5AC region (coded allele G, OR 1·09, 1·06-1·12; p=2·32 × 10-8), and rs560026225 near KIAA1109 (coded allele GATT, OR 1·12, 1·08-1·16; p=3·06 × 10-9). The MUC5AC signal was not associated with asthma when analyses included mild asthma. The rs11603634 G allele was associated with increased expression of MUC5AC mRNA in bronchial epithelial brush samples via proxy SNP rs11602802; (p=2·50 × 10-5) and MUC5AC mRNA was increased in bronchial epithelial samples from patients with severe asthma (in two independent analyses, p=0·039 and p=0·022).
Interpretation: We found substantial shared genetic architecture between mild and moderate-to-severe asthma. We also report for the first time genetic variants associated with the risk of developing moderate-to-severe asthma that regulate mucin production. Finally, we identify candidate causal genes in these loci and provide increased insight into this difficult to treat population.
Funding: Asthma UK, AirPROM, U-BIOPRED, UK Medical Research Council, and Rosetrees Trust.
Copyright © 2019 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Figures



Comment in
-
Genetic insights into moderate-to-severe asthma.Lancet Respir Med. 2019 Jan;7(1):2-3. doi: 10.1016/S2213-2600(18)30447-8. Epub 2018 Dec 11. Lancet Respir Med. 2019. PMID: 30552066 No abstract available.
References
-
- GBD 2015 Chronic Respiratory Disease Collaborators Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. 2017;5:691–706. - PMC - PubMed
-
- Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

