Nucleosome remodeling at origins of global genome-nucleotide excision repair occurs at the boundaries of higher-order chromatin structure
- PMID: 30552104
- PMCID: PMC6314166
- DOI: 10.1101/gr.237198.118
Nucleosome remodeling at origins of global genome-nucleotide excision repair occurs at the boundaries of higher-order chromatin structure
Abstract
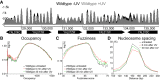
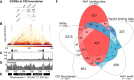
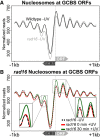
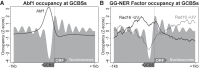
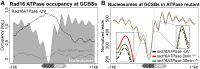
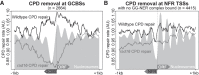
Repair of UV-induced DNA damage requires chromatin remodeling. How repair is initiated in chromatin remains largely unknown. We recently demonstrated that global genome-nucleotide excision repair (GG-NER) in chromatin is organized into domains in relation to open reading frames. Here, we define these domains, identifying the genomic locations from which repair is initiated. By examining DNA damage-induced changes in the linear structure of nucleosomes at these sites, we demonstrate how chromatin remodeling is initiated during GG-NER. In undamaged cells, we show that the GG-NER complex occupies chromatin, establishing the nucleosome structure at these genomic locations, which we refer to as GG-NER complex binding sites (GCBSs). We demonstrate that these sites are frequently located at genomic boundaries that delineate chromosomally interacting domains (CIDs). These boundaries define domains of higher-order nucleosome-nucleosome interaction. We demonstrate that initiation of GG-NER in chromatin is accompanied by the disruption of dynamic nucleosomes that flank GCBSs by the GG-NER complex.
© 2019 van Eijk et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
Global genome nucleotide excision repair is organized into domains that promote efficient DNA repair in chromatin.Genome Res. 2016 Oct;26(10):1376-1387. doi: 10.1101/gr.209106.116. Epub 2016 Jul 28. Genome Res. 2016. PMID: 27470111 Free PMC article.
-
Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin.Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18608-18616. doi: 10.1073/pnas.2003868117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690696 Free PMC article.
-
ABF1-binding sites promote efficient global genome nucleotide excision repair.J Biol Chem. 2009 Jan 9;284(2):966-73. doi: 10.1074/jbc.M806830200. Epub 2008 Nov 6. J Biol Chem. 2009. PMID: 18996839 Free PMC article.
-
Repair of UV lesions in nucleosomes--intrinsic properties and remodeling.DNA Repair (Amst). 2005 Jul 28;4(8):855-69. doi: 10.1016/j.dnarep.2005.04.005. DNA Repair (Amst). 2005. PMID: 15925550 Review.
-
The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair.Int J Mol Sci. 2012;13(9):11954-11973. doi: 10.3390/ijms130911954. Epub 2012 Sep 20. Int J Mol Sci. 2012. PMID: 23109894 Free PMC article. Review.
Cited by
-
GG-NER's role in androgen receptor signaling inhibitor response for advanced prostate cancer.Cell Commun Signal. 2024 Dec 18;22(1):600. doi: 10.1186/s12964-024-01977-0. Cell Commun Signal. 2024. PMID: 39696559 Free PMC article.
-
Set2 histone methyltransferase regulates transcription coupled-nucleotide excision repair in yeast.PLoS Genet. 2022 Mar 9;18(3):e1010085. doi: 10.1371/journal.pgen.1010085. eCollection 2022 Mar. PLoS Genet. 2022. PMID: 35263330 Free PMC article.
-
Nucleotide excision repair leaves a mark on chromatin: DNA damage detection in nucleosomes.Cell Mol Life Sci. 2021 Dec;78(24):7925-7942. doi: 10.1007/s00018-021-03984-7. Epub 2021 Nov 3. Cell Mol Life Sci. 2021. PMID: 34731255 Free PMC article. Review.
-
Super hotspots and super coldspots in the repair of UV-induced DNA damage in the human genome.J Biol Chem. 2021 Jan-Jun;296:100581. doi: 10.1016/j.jbc.2021.100581. Epub 2021 Mar 23. J Biol Chem. 2021. PMID: 33771559 Free PMC article.
-
A quantitative modelling approach for DNA repair on a population scale.PLoS Comput Biol. 2022 Sep 12;18(9):e1010488. doi: 10.1371/journal.pcbi.1010488. eCollection 2022 Sep. PLoS Comput Biol. 2022. PMID: 36094963 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
