Bacillus velezensis Wall Teichoic Acids Are Required for Biofilm Formation and Root Colonization
- PMID: 30552189
- PMCID: PMC6384100
- DOI: 10.1128/AEM.02116-18
Bacillus velezensis Wall Teichoic Acids Are Required for Biofilm Formation and Root Colonization
Abstract
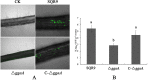
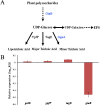
Rhizosphere colonization by plant growth-promoting rhizobacteria (PGPR) along plant roots facilitates the ability of PGPR to promote plant growth and health. Thus, an understanding of the molecular mechanisms of the root colonization process by plant-beneficial Bacillus strains is essential for the use of these strains in agriculture. Here, we observed that an sfp gene mutant of the plant growth-promoting rhizobacterium Bacillus velezensis SQR9 was unable to form normal biofilm architecture, and differential protein expression was observed by proteomic analysis. A minor wall teichoic acid (WTA) biosynthetic protein, GgaA, was decreased over 4-fold in the Δsfp mutant, and impairment of the ggaA gene postponed biofilm formation and decreased cucumber root colonization capabilities. In addition, we provide evidence that the major WTA biosynthetic enzyme GtaB is involved in both biofilm formation and root colonization. The deficiency in biofilm formation of the ΔgtaB mutant may be due to an absence of UDP-glucose, which is necessary for the synthesis of biofilm matrix exopolysaccharides (EPS). These observations provide insights into the root colonization process by a plant-beneficial Bacillus strain, which will help improve its application as a biofertilizer.IMPORTANCEBacillus velezensis is a Gram-positive plant-beneficial bacterium which is widely used in agriculture. Additionally, Bacillus spp. are some of the model organisms used in the study of biofilms, and as such, the molecular networks and regulation systems of biofilm formation are well characterized. However, the molecular processes involved in root colonization by plant-beneficial Bacillus strains remain largely unknown. Here, we showed that WTAs play important roles in the plant root colonization process. The loss of the gtaB gene affects the ability of B. velezensis SQR9 to sense plant polysaccharides, which are important environmental cues that trigger biofilm formation and colonization in the rhizosphere. This knowledge provides new insights into the Bacillus root colonization process and can help improve our understanding of plant-rhizobacterium interactions.
Keywords: Bacillus velezensis SQR9; UDP-glucose; biofilm formation; root colonization; wall teichoic acids.
Copyright © 2019 American Society for Microbiology.
Figures




Similar articles
-
FtsEX-CwlO regulates biofilm formation by a plant-beneficial rhizobacterium Bacillus velezensis SQR9.Res Microbiol. 2018 Apr;169(3):166-176. doi: 10.1016/j.resmic.2018.01.004. Epub 2018 Feb 8. Res Microbiol. 2018. PMID: 29427638
-
Quorum sensing signal autoinducer-2 promotes root colonization of Bacillus velezensis SQR9 by affecting biofilm formation and motility.Appl Microbiol Biotechnol. 2020 Aug;104(16):7177-7185. doi: 10.1007/s00253-020-10713-w. Epub 2020 Jul 4. Appl Microbiol Biotechnol. 2020. PMID: 32621125
-
Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection.FEMS Microbiol Lett. 2014 Apr;353(1):49-56. doi: 10.1111/1574-6968.12406. Epub 2014 Mar 19. FEMS Microbiol Lett. 2014. PMID: 24612247
-
Bacterial cell differentiation during plant root colonization: the putative role of fructans.Physiol Plant. 2025 Jan-Feb;177(1):e70095. doi: 10.1111/ppl.70095. Physiol Plant. 2025. PMID: 39887703 Review.
-
Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance.J Basic Microbiol. 2018 Dec;58(12):1009-1022. doi: 10.1002/jobm.201800309. Epub 2018 Sep 5. J Basic Microbiol. 2018. PMID: 30183106 Review.
Cited by
-
The complete genome sequences of Bacillus velezensis B26: a promising biocontrol agent and biofertilizer.F1000Res. 2025 Jul 8;14:170. doi: 10.12688/f1000research.160546.2. eCollection 2025. F1000Res. 2025. PMID: 40495924 Free PMC article.
-
A phosphate starvation induced small RNA promotes Bacillus biofilm formation.NPJ Biofilms Microbiomes. 2024 Oct 29;10(1):115. doi: 10.1038/s41522-024-00586-6. NPJ Biofilms Microbiomes. 2024. PMID: 39472585 Free PMC article.
-
Plant growth-promoting activities and genomic analysis of the stress-resistant Bacillus megaterium STB1, a bacterium of agricultural and biotechnological interest.Biotechnol Rep (Amst). 2019 Dec 4;25:e00406. doi: 10.1016/j.btre.2019.e00406. eCollection 2020 Mar. Biotechnol Rep (Amst). 2019. PMID: 31886139 Free PMC article.
-
Biofertilizer Industry and Research Developments in China: A Mini-Review.Microb Biotechnol. 2025 May;18(5):e70163. doi: 10.1111/1751-7915.70163. Microb Biotechnol. 2025. PMID: 40411486 Free PMC article. Review.
-
Amino Acids From Root Exudates Induce Bacillus Spore Germination to Enhance Root Colonisation and Plant Growth Promotion.Microb Biotechnol. 2025 Jun;18(6):e70172. doi: 10.1111/1751-7915.70172. Microb Biotechnol. 2025. PMID: 40448297 Free PMC article.
References
-
- Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Süssmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R. 2007. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25:1007–1014. doi:10.1038/nbt1325. - DOI - PubMed
-
- Chet I, Chernin L. 2003. Biocontrol, microbial agents in soil, p 450–465. In Bitton G. (ed), Encyclopedia of environmental microbiology. John Wiley & Sons, New York, NY.
-
- Abed H, Rouag N, Mouatassem D, Rouabhi A. 2016. Screening for Pseudomonas and Bacillus antagonistic rhizobacteria strains for the biocontrol of Fusarium wilt of chickpea. Eurasian J Soil Sci 5:182–191. doi:10.18393/ejss.2016.3.182-191. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

