Contrasting Patterns of Virus Protection and Functional Incompatibility Genes in Two Conspecific Wolbachia Strains from Drosophila pandora
- PMID: 30552191
- PMCID: PMC6384105
- DOI: 10.1128/AEM.02290-18
Contrasting Patterns of Virus Protection and Functional Incompatibility Genes in Two Conspecific Wolbachia Strains from Drosophila pandora
Abstract
Wolbachia infections can present different phenotypes in hosts, including different forms of reproductive manipulation and antiviral protection, which may influence infection dynamics within host populations. In populations of Drosophila pandora two distinct Wolbachia strains coexist, each manipulating host reproduction: strain wPanCI causes cytoplasmic incompatibility (CI), whereas strain wPanMK causes male killing (MK). CI occurs when a Wolbachia-infected male mates with a female not infected with a compatible type of Wolbachia, leading to nonviable offspring. wPanMK can rescue wPanCI-induced CI but is unable to induce CI. The antiviral protection phenotypes provided by the wPanCI and wPanMK infections were characterized; the strains showed differential protection phenotypes, whereby cricket paralysis virus (CrPV)-induced mortality was delayed in flies infected with wPanMK but enhanced in flies infected with wPanCI compared to their respective Wolbachia-cured counterparts. Homologs of the cifA and cifB genes involved in CI identified in wPanMK and wPanCI showed a high degree of conservation; however, the CifB protein in wPanMK is truncated and is likely nonfunctional. The presence of a likely functional CifA in wPanMK and wPanMK's ability to rescue wPanCI-induced CI are consistent with the recent confirmation of CifA's involvement in CI rescue, and the absence of a functional CifB protein further supports its involvement as a CI modification factor. Taken together, these findings indicate that wPanCI and wPanMK have different relationships with their hosts in terms of their protective and CI phenotypes. It is therefore likely that different factors influence the prevalence and dynamics of these coinfections in natural Drosophila pandora hosts.IMPORTANCEWolbachia strains are common endosymbionts in insects, with multiple strains often coexisting in the same species. The coexistence of multiple strains is poorly understood but may rely on Wolbachia organisms having diverse phenotypic effects on their hosts. As Wolbachia is increasingly being developed as a tool to control disease transmission and suppress pest populations, it is important to understand the ways in which multiple Wolbachia strains persist in natural populations and how these might then be manipulated. We have therefore investigated viral protection and the molecular basis of cytoplasmic incompatibility in two coexisting Wolbachia strains with contrasting effects on host reproduction.
Keywords: Dicistroviridae; Wolbachia; antiviral protection; cytoplasmic incompatibility; male killing; virus blocking.
Copyright © 2019 American Society for Microbiology.
Figures
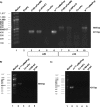
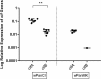
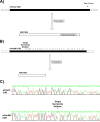


Similar articles
-
Evolutionary Genetics of Cytoplasmic Incompatibility Genes cifA and cifB in Prophage WO of Wolbachia.Genome Biol Evol. 2018 Feb 1;10(2):434-451. doi: 10.1093/gbe/evy012. Genome Biol Evol. 2018. PMID: 29351633 Free PMC article.
-
Parallel Sequencing of Wolbachia wCer2 from Donor and Novel Hosts Reveals Multiple Incompatibility Factors and Genome Stability after Host Transfers.Genome Biol Evol. 2020 May 1;12(5):720-735. doi: 10.1093/gbe/evaa050. Genome Biol Evol. 2020. PMID: 32163151 Free PMC article.
-
The impacts of cytoplasmic incompatibility factor (cifA and cifB) genetic variation on phenotypes.Genetics. 2021 Mar 3;217(1):1-13. doi: 10.1093/genetics/iyaa007. Genetics. 2021. PMID: 33683351 Free PMC article.
-
Sperm chromatin remodelling and Wolbachia-induced cytoplasmic incompatibility in Drosophila.Biochem Cell Biol. 2003 Jun;81(3):229-40. doi: 10.1139/o03-053. Biochem Cell Biol. 2003. PMID: 12897857 Review.
-
The Biochemistry of Cytoplasmic Incompatibility Caused by Endosymbiotic Bacteria.Genes (Basel). 2020 Jul 25;11(8):852. doi: 10.3390/genes11080852. Genes (Basel). 2020. PMID: 32722516 Free PMC article. Review.
Cited by
-
Genetic innovations in animal-microbe symbioses.Nat Rev Genet. 2022 Jan;23(1):23-39. doi: 10.1038/s41576-021-00395-z. Epub 2021 Aug 13. Nat Rev Genet. 2022. PMID: 34389828 Free PMC article. Review.
-
Wolbachia-Virus interactions and arbovirus control through population replacement in mosquitoes.Pathog Glob Health. 2023 May;117(3):245-258. doi: 10.1080/20477724.2022.2117939. Epub 2022 Oct 7. Pathog Glob Health. 2023. PMID: 36205550 Free PMC article.
-
Symbiont-mediated cytoplasmic incompatibility: what have we learned in 50 years?Elife. 2020 Sep 25;9:e61989. doi: 10.7554/eLife.61989. Elife. 2020. PMID: 32975515 Free PMC article. Review.
-
Life and Death of Selfish Genes: Comparative Genomics Reveals the Dynamic Evolution of Cytoplasmic Incompatibility.Mol Biol Evol. 2021 Jan 4;38(1):2-15. doi: 10.1093/molbev/msaa209. Mol Biol Evol. 2021. PMID: 32797213 Free PMC article.
-
Wolbachia strain wAlbB maintains high density and dengue inhibition following introduction into a field population of Aedes aegypti.Philos Trans R Soc Lond B Biol Sci. 2021 Feb 15;376(1818):20190809. doi: 10.1098/rstb.2019.0809. Epub 2020 Dec 28. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33357050 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

