Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9
- PMID: 30552336
- PMCID: PMC6293998
- DOI: 10.1038/s41467-018-07784-9
Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9
Erratum in
-
Author Correction: Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9.Nat Commun. 2019 Jul 23;10(1):3351. doi: 10.1038/s41467-019-11310-w. Nat Commun. 2019. PMID: 31337757 Free PMC article.
Abstract
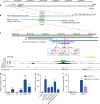
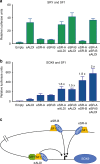
Disorders of sex development (DSDs) are conditions affecting development of the gonads or genitalia. Variants in two key genes, SRY and its target SOX9, are an established cause of 46,XY DSD, but the genetic basis of many DSDs remains unknown. SRY-mediated SOX9 upregulation in the early gonad is crucial for testis development, yet the regulatory elements underlying this have not been identified in humans. Here, we identified four DSD patients with overlapping duplications or deletions upstream of SOX9. Bioinformatic analysis identified three putative enhancers for SOX9 that responded to different combinations of testis-specific regulators. All three enhancers showed synergistic activity and together drive SOX9 in the testis. This is the first study to identify SOX9 enhancers that, when duplicated or deleted, result in 46,XX or 46,XY sex reversal, respectively. These enhancers provide a hitherto missing link by which SRY activates SOX9 in humans, and establish SOX9 enhancer mutations as a significant cause of DSD.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

