Skewed X-inactivation is common in the general female population
- PMID: 30552425
- PMCID: PMC6460563
- DOI: 10.1038/s41431-018-0291-3
Skewed X-inactivation is common in the general female population
Abstract
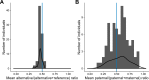
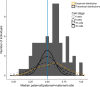
X-inactivation is a well-established dosage compensation mechanism ensuring that X-chromosomal genes are expressed at comparable levels in males and females. Skewed X-inactivation is often explained by negative selection of one of the alleles. We demonstrate that imbalanced expression of the paternal and maternal X-chromosomes is common in the general population and that the random nature of the X-inactivation mechanism can be sufficient to explain the imbalance. To this end, we analyzed blood-derived RNA and whole-genome sequencing data from 79 female children and their parents from the Genome of the Netherlands project. We calculated the median ratio of the paternal over total counts at all X-chromosomal heterozygous single-nucleotide variants with coverage ≥10. We identified two individuals where the same X-chromosome was inactivated in all cells. Imbalanced expression of the two X-chromosomes (ratios ≤0.35 or ≥0.65) was observed in nearly 50% of the population. The empirically observed skewing is explained by a theoretical model where X-inactivation takes place in an embryonic stage in which eight cells give rise to the hematopoietic compartment. Genes escaping X-inactivation are expressed from both alleles and therefore demonstrate less skewing than inactivated genes. Using this characteristic, we identified three novel escapee genes (SSR4, REPS2, and SEPT6), but did not find support for many previously reported escapee genes in blood. Our collective data suggest that skewed X-inactivation is common in the general population. This may contribute to manifestation of symptoms in carriers of recessive X-linked disorders. We recommend that X-inactivation results should not be used lightly in the interpretation of X-linked variants.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

