Live imaging reveals hub cell assembly and compaction dynamics during morphogenesis of the Drosophila testis niche
- PMID: 30553808
- PMCID: PMC6410746
- DOI: 10.1016/j.ydbio.2018.12.014
Live imaging reveals hub cell assembly and compaction dynamics during morphogenesis of the Drosophila testis niche
Abstract
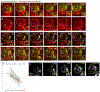
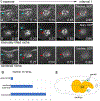
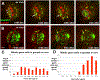
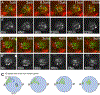
Adult stem cells are often found in specialized niches, where the constituent cells direct self-renewal of their stem cell pool. The niche is therefore crucial for both normal homeostasis and tissue regeneration. In many mammalian tissues, niche cells have classically been difficult to identify, which has hampered any understanding of how tissues first construct niches during development. Fortunately, the Drosophila germline stem cell (GSC) niche is well defined, allowing for unambiguous identification of both niche cells and resident stem cells. The testis niche first forms in the early embryo, during a late stage of gonadogenesis. Here, using live-imaging both in vivo and ex vivo, we follow pro-niche cells as they assemble and assume their final form. We show that after ex vivo culture the niche appears fully functional, as judged by enrichment of adhesion proteins, the ability to activate STAT in adjacent GSCs, and to direct GSCs to divide orthogonally to the niche, just as they would in situ. Collectively, our imaging has generated several novel insights on niche morphogenesis that could not be inferred from fixed images alone. We identify dynamic processes that constitute an assembly phase and a compaction phase during morphogenesis. The compaction phase correlates with cell neighbor exchange among the assembled pro-niche cells, as well as a burst of divisions among newly recruited stem cells. Before compaction, an assembly phase involves the movement of pro-niche cells along the outer periphery of the gonad, using the extracellular matrix (ECM) to assemble at the anterior of the gonad. Finally, live-imaging in integrin mutants allows us to define the role of pro-niche cell-ECM interaction with regard to the new assembly and compaction dynamics revealed here.
Keywords: Drosophila testis stem cell niche; Gonadogenesis; Niche morphogenesis.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures




References
-
- Calvi LM et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425, 841–846 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

