Nucleosome Positioning by an Evolutionarily Conserved Chromatin Remodeler Prevents Aberrant DNA Methylation in Neurospora
- PMID: 30554169
- PMCID: PMC6366918
- DOI: 10.1534/genetics.118.301711
Nucleosome Positioning by an Evolutionarily Conserved Chromatin Remodeler Prevents Aberrant DNA Methylation in Neurospora
Abstract
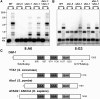
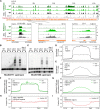
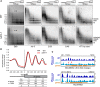
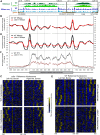
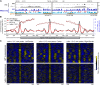
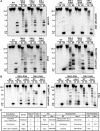
In the filamentous fungus Neurospora crassa, constitutive heterochromatin is marked by tri-methylation of histone H3 lysine 9 (H3K9me3) and DNA methylation. We identified mutations in the Neurospora defective in methylation-1 (dim-1) gene that cause defects in cytosine methylation and implicate a putative AAA-ATPase chromatin remodeler. Although it was well-established that chromatin remodelers can affect transcription by influencing DNA accessibility with nucleosomes, little was known about the role of remodelers on chromatin that is normally not transcribed, including regions of constitutive heterochromatin. We found that dim-1 mutants display both reduced DNA methylation in heterochromatic regions as well as increased DNA methylation and H3K9me3 in some intergenic regions associated with highly expressed genes. Deletion of dim-1 leads to atypically spaced nucleosomes throughout the genome and numerous changes in gene expression. DIM-1 localizes to both heterochromatin and intergenic regions that become hyper-methylated in dim-1 strains. Our findings indicate that DIM-1 normally positions nucleosomes in both heterochromatin and euchromatin and that the standard arrangement and density of nucleosomes is required for the proper function of heterochromatin machinery.
Keywords: CATP; DIM-1; DNA methylation; Neurospora crassa; heterochromatin; nucleosome.
Copyright © 2019 by the Genetics Society of America.
Figures
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

