Repression of Transcription at DNA Breaks Requires Cohesin throughout Interphase and Prevents Genome Instability
- PMID: 30554942
- PMCID: PMC6344341
- DOI: 10.1016/j.molcel.2018.11.001
Repression of Transcription at DNA Breaks Requires Cohesin throughout Interphase and Prevents Genome Instability
Abstract
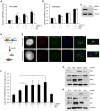
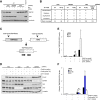
Cohesin subunits are frequently mutated in cancer, but how they function as tumor suppressors is unknown. Cohesin mediates sister chromatid cohesion, but this is not always perturbed in cancer cells. Here, we identify a previously unknown role for cohesin. We find that cohesin is required to repress transcription at DNA double-strand breaks (DSBs). Notably, cohesin represses transcription at DSBs throughout interphase, indicating that this is distinct from its known role in mediating DNA repair through sister chromatid cohesion. We identified a cancer-associated SA2 mutation that supports sister chromatid cohesion but is unable to repress transcription at DSBs. We further show that failure to repress transcription at DSBs leads to large-scale genome rearrangements. Cancer samples lacking SA2 display mutational patterns consistent with loss of this pathway. These findings uncover a new function for cohesin that provides insights into its frequent loss in cancer.
Keywords: DNA repair; PBAF; PBRM1; SA2; SMARCA4; STAG2; SWI/SNF; cancer; transcriptional silencing.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

