Quantitative RNAseq analysis of Ugandan KS tumors reveals KSHV gene expression dominated by transcription from the LTd downstream latency promoter
- PMID: 30557332
- PMCID: PMC6312348
- DOI: 10.1371/journal.ppat.1007441
Quantitative RNAseq analysis of Ugandan KS tumors reveals KSHV gene expression dominated by transcription from the LTd downstream latency promoter
Abstract
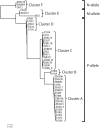
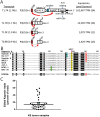
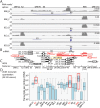
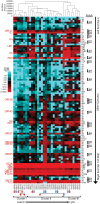
KSHV is endemic in Uganda and the HIV epidemic has dramatically increased the incidence of Kaposi sarcoma (KS). To investigate the role of KSHV in the development of KS, we obtained KS biopsies from ART-naïve, HIV-positive individuals in Uganda and analyzed the tumors using RNAseq to globally characterize the KSHV transcriptome. Phylogenetic analysis of ORF75 sequences from 23 tumors revealed 6 distinct genetic clusters with KSHV strains exhibiting M, N or P alleles. RNA reads mapping to specific unique coding sequence (UCDS) features were quantitated using a gene feature file previously developed to globally analyze and quantitate KSHV transcription in infected endothelial cells. A pattern of high level expression was detected in the KSHV latency region that was common to all KS tumors. The clear majority of transcription was derived from the downstream latency transcript promoter P3(LTd) flanking ORF72, with little evidence of transcription from the P1(LTc) latency promoter, which is constitutive in KSHV-infected lymphomas and tissue-culture cells. RNAseq data provided evidence of alternate P3(LTd) transcript editing, splicing and termination resulting in multiple gene products, with 90% of the P3(LTd) transcripts spliced to release the intronic source of the microRNAs K1-9 and 11. The spliced transcripts encode a regulatory uORF upstream of Kaposin A with alterations in intervening repeat sequences yielding novel or deleted Kaposin B/C-like sequences. Hierarchical clustering and PCA analysis of KSHV transcripts revealed three clusters of tumors with different latent and lytic gene expression profiles. Paradoxically, tumors with a latent phenotype had high levels of total KSHV transcription, while tumors with a lytic phenotype had low levels of total KSHV transcription. Morphologically distinct KS tumors from the same individual showed similar KSHV gene expression profiles suggesting that the tumor microenvironment and host response play important roles in the activation level of KSHV within the infected tumor cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
Kaposi's Sarcoma-Associated Herpesvirus Infection Induces the Expression of Neuroendocrine Genes in Endothelial Cells.J Virol. 2020 Mar 31;94(8):e01692-19. doi: 10.1128/JVI.01692-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31969437 Free PMC article.
-
The elevated expression of ORF75, a KSHV lytic gene, in Kaposi sarcoma lesions is driven by a GC-rich DNA cis element in its promoter region.PLoS Pathog. 2025 Mar 17;21(3):e1012984. doi: 10.1371/journal.ppat.1012984. eCollection 2025 Mar. PLoS Pathog. 2025. PMID: 40096169 Free PMC article.
-
A Panel of Kaposi's Sarcoma-Associated Herpesvirus Mutants in the Polycistronic Kaposin Locus for Precise Analysis of Individual Protein Products.J Virol. 2022 Mar 9;96(5):e0156021. doi: 10.1128/JVI.01560-21. Epub 2021 Dec 22. J Virol. 2022. PMID: 34936820 Free PMC article.
-
KSHV Genome Replication and Maintenance in Latency.Adv Exp Med Biol. 2018;1045:299-320. doi: 10.1007/978-981-10-7230-7_14. Adv Exp Med Biol. 2018. PMID: 29896673 Review.
-
Endothelial cell- and lymphocyte-based in vitro systems for understanding KSHV biology.Curr Top Microbiol Immunol. 2007;312:211-44. doi: 10.1007/978-3-540-34344-8_8. Curr Top Microbiol Immunol. 2007. PMID: 17089799 Review.
Cited by
-
Single-cell analysis of Kaposi's sarcoma-associated herpesvirus infection in three-dimensional air-liquid interface culture model.PLoS Pathog. 2022 Aug 17;18(8):e1010775. doi: 10.1371/journal.ppat.1010775. eCollection 2022 Aug. PLoS Pathog. 2022. PMID: 35976902 Free PMC article.
-
Variation within major internal repeats of KSHV in vivo.Virus Evol. 2023 May 22;9(1):vead034. doi: 10.1093/ve/vead034. eCollection 2023. Virus Evol. 2023. PMID: 37325087 Free PMC article.
-
Intra-host changes in Kaposi sarcoma-associated herpesvirus genomes in Ugandan adults with Kaposi sarcoma.PLoS Pathog. 2021 Jan 19;17(1):e1008594. doi: 10.1371/journal.ppat.1008594. eCollection 2021 Jan. PLoS Pathog. 2021. PMID: 33465147 Free PMC article.
-
T-cells specific for KSHV and HIV migrate to Kaposi sarcoma tumors and persist over time.bioRxiv [Preprint]. 2025 Feb 15:2024.02.06.579223. doi: 10.1101/2024.02.06.579223. bioRxiv. 2025. PMID: 38370623 Free PMC article. Preprint.
-
Comparative transcriptome analysis of endemic and epidemic Kaposi's sarcoma (KS) lesions and the secondary role of HIV-1 in KS pathogenesis.PLoS Pathog. 2020 Jul 24;16(7):e1008681. doi: 10.1371/journal.ppat.1008681. eCollection 2020 Jul. PLoS Pathog. 2020. PMID: 32706839 Free PMC article.
References
-
- Nicholas J, Zong JC, Alcendor DJ, Ciufo DM, Poole LJ, Sarisky RT, et al. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;(23):79–88. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

