Evolutionary Analysis of Plastid Genomes of Seven Lonicera L. Species: Implications for Sequence Divergence and Phylogenetic Relationships
- PMID: 30558106
- PMCID: PMC6321470
- DOI: 10.3390/ijms19124039
Evolutionary Analysis of Plastid Genomes of Seven Lonicera L. Species: Implications for Sequence Divergence and Phylogenetic Relationships
Abstract
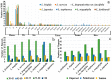
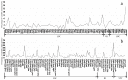
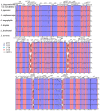
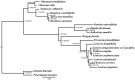
Plant plastomes play crucial roles in species evolution and phylogenetic reconstruction studies due to being maternally inherited and due to the moderate evolutionary rate of genomes. However, patterns of sequence divergence and molecular evolution of the plastid genomes in the horticulturally- and economically-important Lonicera L. species are poorly understood. In this study, we collected the complete plastomes of seven Lonicera species and determined the various repeat sequence variations and protein sequence evolution by comparative genomic analysis. A total of 498 repeats were identified in plastid genomes, which included tandem (130), dispersed (277), and palindromic (91) types of repeat variations. Simple sequence repeat (SSR) elements analysis indicated the enriched SSRs in seven genomes to be mononucleotides, followed by tetra-nucleotides, dinucleotides, tri-nucleotides, hex-nucleotides, and penta-nucleotides. We identified 18 divergence hotspot regions (rps15, rps16, rps18, rpl23, psaJ, infA, ycf1, trnN-GUU-ndhF, rpoC2-rpoC1, rbcL-psaI, trnI-CAU-ycf2, psbZ-trnG-UCC, trnK-UUU-rps16, infA-rps8, rpl14-rpl16, trnV-GAC-rrn16, trnL-UAA intron, and rps12-clpP) that could be used as the potential molecular genetic markers for the further study of population genetics and phylogenetic evolution of Lonicera species. We found that a large number of repeat sequences were distributed in the divergence hotspots of plastid genomes. Interestingly, 16 genes were determined under positive selection, which included four genes for the subunits of ribosome proteins (rps7, rpl2, rpl16, and rpl22), three genes for the subunits of photosystem proteins (psaJ, psbC, and ycf4), three NADH oxidoreductase genes (ndhB, ndhH, and ndhK), two subunits of ATP genes (atpA and atpB), and four other genes (infA, rbcL, ycf1, and ycf2). Phylogenetic analysis based on the whole plastome demonstrated that the seven Lonicera species form a highly-supported monophyletic clade. The availability of these plastid genomes provides important genetic information for further species identification and biological research on Lonicera.
Keywords: Lonicera; phylogenetic relationship; plastid genome; positive selection; repeat sequences.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Comparative Chloroplast Genomics of Dipsacales Species: Insights Into Sequence Variation, Adaptive Evolution, and Phylogenetic Relationships.Front Plant Sci. 2018 May 23;9:689. doi: 10.3389/fpls.2018.00689. eCollection 2018. Front Plant Sci. 2018. PMID: 29875791 Free PMC article.
-
Characterization of the complete chloroplast genome sequences of six Dalbergia species and its comparative analysis in the subfamily of Papilionoideae (Fabaceae).PeerJ. 2022 Jul 1;10:e13570. doi: 10.7717/peerj.13570. eCollection 2022. PeerJ. 2022. PMID: 35795179 Free PMC article.
-
Molecular Evolution of Chloroplast Genomes of Orchid Species: Insights into Phylogenetic Relationship and Adaptive Evolution.Int J Mol Sci. 2018 Mar 2;19(3):716. doi: 10.3390/ijms19030716. Int J Mol Sci. 2018. PMID: 29498674 Free PMC article.
-
Dynamic evolution of the plastome in the Elm family (Ulmaceae).Planta. 2022 Dec 17;257(1):14. doi: 10.1007/s00425-022-04045-4. Planta. 2022. PMID: 36526857 Review.
-
Small Genomes and Big Data: Adaptation of Plastid Genomics to the High-Throughput Era.Biomolecules. 2019 Jul 24;9(8):299. doi: 10.3390/biom9080299. Biomolecules. 2019. PMID: 31344945 Free PMC article. Review.
Cited by
-
Chloroplast genomes of Simarouba Aubl., molecular evolution and comparative analyses within Sapindales.Sci Rep. 2024 Sep 12;14(1):21358. doi: 10.1038/s41598-024-71956-5. Sci Rep. 2024. PMID: 39266625 Free PMC article.
-
Plastome phylogenomics and morphological traits analyses provide new insights into the phylogenetic position, species delimitation and speciation of Triplostegia (Caprifoliaceae).BMC Plant Biol. 2023 Dec 15;23(1):645. doi: 10.1186/s12870-023-04663-4. BMC Plant Biol. 2023. PMID: 38097946 Free PMC article.
-
Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation.Int J Mol Sci. 2024 Feb 12;25(4):2193. doi: 10.3390/ijms25042193. Int J Mol Sci. 2024. PMID: 38396871 Free PMC article.
-
The first complete chloroplast genome of Thalictrum fargesii: insights into phylogeny and species identification.Front Plant Sci. 2024 Apr 29;15:1356912. doi: 10.3389/fpls.2024.1356912. eCollection 2024. Front Plant Sci. 2024. PMID: 38745930 Free PMC article.
-
Complete Chloroplast Genome Sequence of Triosteum sinuatum, Insights into Comparative Chloroplast Genomics, Divergence Time Estimation and Phylogenetic Relationships among Dipsacales.Genes (Basel). 2022 May 23;13(5):933. doi: 10.3390/genes13050933. Genes (Basel). 2022. PMID: 35627318 Free PMC article.
References
-
- Hsu P.S. A preliminary numerical taxonomy of the family Caprifoliaceae. Acta Phytotaxon. Sin. 1983;21:26–32.
-
- Li H.J., Li P., Wang M.C., Ye W.C. A new secoiridoid glucoside from Lonicera japonica. Chin. J. Nat. Med. 2003;3:132–133.
-
- Ren M.T., Chen J., Song Y., Sheng L.S., Li P., Qi L.W. Identification and quantification of 32 bioactive compounds in Lonicera species by high performance liquid chromatography coupled with time-of-flight mass spectrometry. J. Pharm. Biomed. 2008;48:1351–1360. doi: 10.1016/j.jpba.2008.09.037. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

