Simultaneous Detection of Different Zika Virus Lineages via Molecular Computation in a Point-of-Care Assay
- PMID: 30558136
- PMCID: PMC6316447
- DOI: 10.3390/v10120714
Simultaneous Detection of Different Zika Virus Lineages via Molecular Computation in a Point-of-Care Assay
Abstract
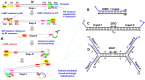
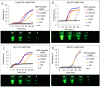
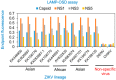
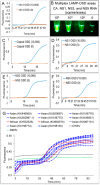
We have developed a generalizable "smart molecular diagnostic" capable of accurate point-of-care (POC) detection of variable nucleic acid targets. Our isothermal assay relies on multiplex execution of four loop-mediated isothermal amplification reactions, with primers that are degenerate and redundant, thereby increasing the breadth of targets while reducing the probability of amplification failure. An easy-to-read visual answer is computed directly by a multi-input Boolean OR logic gate (gate output is true if either one or more gate inputs is true) signal transducer that uses degenerate strand exchange probes to assess any combination of amplicons. We demonstrate our methodology by using the same assay to detect divergent Asian and African lineages of the evolving Zika virus (ZIKV), while maintaining selectivity against non-target viruses. Direct analysis of biological specimens proved possible, with crudely macerated ZIKV-infected Aedes aegypti mosquitoes being identified with 100% specificity and sensitivity. The ease-of-use with minimal instrumentation, broad programmability, and built-in fail-safe reliability make our smart molecular diagnostic attractive for POC use.
Keywords: boolean logic-processing nucleic acid probes; isothermal nucleic acid amplification; mosquito; mosquito surveillance; multiplex nucleic acid detection; nucleic acid computation; nucleic acid strand exchange; point-of-care diagnostics; zika virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Development and Validation of Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) for Rapid Detection of ZIKV in Mosquito Samples from Brazil.Sci Rep. 2019 Mar 14;9(1):4494. doi: 10.1038/s41598-019-40960-5. Sci Rep. 2019. PMID: 30872672 Free PMC article.
-
Detection of Zika Virus Using Reverse Transcription-Loop-Mediated Isothermal Amplification (RT-LAMP).Methods Mol Biol. 2020;2142:137-146. doi: 10.1007/978-1-0716-0581-3_12. Methods Mol Biol. 2020. PMID: 32367365
-
Lab-on-a-Chip Zika Detection With Reverse Transcription Loop-Mediated Isothermal Amplification-Based Assay for Point-of-Care Settings.Arch Pathol Lab Med. 2020 Nov 1;144(11):1335-1343. doi: 10.5858/arpa.2019-0667-OA. Arch Pathol Lab Med. 2020. PMID: 32886758
-
Loop-Mediated Isothermal Amplification (LAMP) for the Diagnosis of Zika Virus: A Review.Viruses. 2019 Dec 23;12(1):19. doi: 10.3390/v12010019. Viruses. 2019. PMID: 31877989 Free PMC article. Review.
-
Zika virus diagnosis: challenges and solutions.Clin Microbiol Infect. 2019 Feb;25(2):142-146. doi: 10.1016/j.cmi.2018.12.002. Epub 2018 Dec 12. Clin Microbiol Infect. 2019. PMID: 30553031 Review.
Cited by
-
Single-cell nucleic acid profiling in droplets (SNAPD) enables high-throughput analysis of heterogeneous cell populations.Nucleic Acids Res. 2021 Oct 11;49(18):e103. doi: 10.1093/nar/gkab577. Nucleic Acids Res. 2021. PMID: 34233007 Free PMC article.
-
Development of an antibody-dependent cellular cytotoxicity reporter assay for measuring anti-Middle East Respiratory Syndrome antibody bioactivity.Sci Rep. 2020 Oct 6;10(1):16615. doi: 10.1038/s41598-020-73960-x. Sci Rep. 2020. PMID: 33024203 Free PMC article.
-
Developments in integrating nucleic acid isothermal amplification and detection systems for point-of-care diagnostics.Biosens Bioelectron. 2020 Dec 15;170:112674. doi: 10.1016/j.bios.2020.112674. Epub 2020 Oct 2. Biosens Bioelectron. 2020. PMID: 33035900 Free PMC article. Review.
-
Preparation and Use of Cellular Reagents: A Low-resource Molecular Biology Reagent Platform.Curr Protoc. 2022 Mar;2(3):e387. doi: 10.1002/cpz1.387. Curr Protoc. 2022. PMID: 35263038 Free PMC article.
-
Analysis of macerated ticks using Boolean logic gating colorimetric isothermal nucleic acid assays for Lyme Borrelia and Ixodes scapularis ticks.Sci Rep. 2023 Jul 15;13(1):11439. doi: 10.1038/s41598-023-38452-8. Sci Rep. 2023. PMID: 37454160 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

