Label-Free SERS Discrimination and In Situ Analysis of Life Cycle in Escherichia coli and Staphylococcus epidermidis
- PMID: 30558342
- PMCID: PMC6315751
- DOI: 10.3390/bios8040131
Label-Free SERS Discrimination and In Situ Analysis of Life Cycle in Escherichia coli and Staphylococcus epidermidis
Abstract
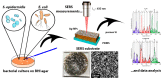
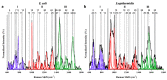
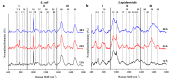
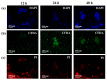
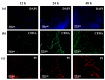
Surface enhanced Raman spectroscopy (SERS) has been proven suitable for identifying and characterizing different bacterial species, and to fully understand the chemically driven metabolic variations that occur during their evolution. In this study, SERS was exploited to identify the cellular composition of Gram-positive and Gram-negative bacteria by using mesoporous silicon-based substrates decorated with silver nanoparticles. The main differences between the investigated bacterial strains reside in the structure of the cell walls and plasmatic membranes, as well as their biofilm matrix, as clearly noticed in the corresponding SERS spectrum. A complete characterization of the spectra was provided in order to understand the contribution of each vibrational signal collected from the bacterial culture at different times, allowing the analysis of the bacterial populations after 12, 24, and 48 h. The results show clear features in terms of vibrational bands in line with the bacterial growth curve, including an increasing intensity of the signals during the first 24 h and their subsequent decrease in the late stationary phase after 48 h of culture. The evolution of the bacterial culture was also confirmed by fluorescence microscope images.
Keywords: E. coli; S. epidermidis; SERS; biofilm; metal-dielectric nanostructures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Novara C., Chiadò A., Paccotti N., Catuogno S., Esposito C.L., Condorelli G., De Franciscis V., Geobaldo F., Rivolo P., Giorgis F. SERS-active metal-dielectric nanostructures integrated in microfluidic devices for label-free quantitative detection of miRNA. Faraday Discuss. 2017;205:271–289. doi: 10.1039/C7FD00140A. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

