Natural APOBEC3C variants can elicit differential HIV-1 restriction activity
- PMID: 30558640
- PMCID: PMC6297987
- DOI: 10.1186/s12977-018-0459-5
Natural APOBEC3C variants can elicit differential HIV-1 restriction activity
Abstract
Background: The APOBEC3 (A3) family of DNA cytosine deaminases provides an innate barrier to infection by retroviruses including HIV-1. A total of five enzymes, A3C, A3D, A3F, A3G and A3H, are degraded by the viral accessory protein Vif and expressed at high levels in CD4+ T cells, the primary reservoir for HIV-1 replication in vivo. Apart from A3C, all of these enzymes mediate restriction of Vif-deficient HIV-1. However, a rare variant of human A3C (Ile188) was shown recently to restrict Vif-deficient HIV-1 in a 293T-based single cycle infection system. The potential activity of this naturally occurring A3C variant has yet to be characterized in a T cell-based spreading infection system. Here we employ a combination of Cas9/gRNA disruption and transient and stable protein expression to assess the roles of major Ser188 and minor Ile188 A3C variants in HIV-1 restriction in T cell lines.
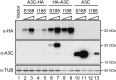
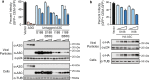
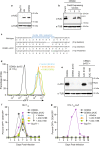
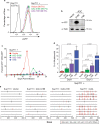
Results: Cas9-mediated mutation of endogenous A3C in the non-permissive CEM2n T cell line did not alter HIV-1 replication kinetics, and complementation with A3C-Ser188 or A3C-Ile188 was similarly aphenotypic. Stable expression of A3C-Ser188 in the permissive T cell line SupT11 also had little effect. However, stable expression of A3C-Ile188 in SupT11 cells inhibited Vif-deficient virus replication and inflicted G-to-A mutations.
Conclusions: A3C-Ile188 is capable of inhibiting Vif-deficient HIV-1 replication in T cells. Although A3C is eclipsed by the dominant anti-viral activities of other A3s in non-permissive T cell lines and primary T lymphocytes, this enzyme may still be able to contribute to HIV-1 diversification in vivo. Our results highlight the functional redundancy in the human A3 family with regards to HIV-1 restriction and the need to consider naturally occurring variants.
Keywords: APOBEC3C; DNA cytosine deaminase; HIV-1; Human genetic variation; Innate immunity; Retrovirus restriction factor.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

