Conformations of an RNA Helix-Junction-Helix Construct Revealed by SAXS Refinement of MD Simulations
- PMID: 30558889
- PMCID: PMC6341278
- DOI: 10.1016/j.bpj.2018.11.020
Conformations of an RNA Helix-Junction-Helix Construct Revealed by SAXS Refinement of MD Simulations
Abstract
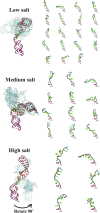
RNA is involved in a broad range of biological processes that extend far beyond translation. Many of RNA's recently discovered functions rely on folding to a specific conformation or transitioning between conformations. The RNA structure contains rigid, short basepaired regions connected by more flexible linkers. Studies of model constructs such as small helix-junction-helix (HJH) motifs are useful in understanding how these elements work together to determine RNA conformation. Here, we reveal the full ensemble of solution structures assumed by a model RNA HJH. We apply small-angle x-ray scattering and an ensemble optimization method to selectively refine models generated by all-atom molecular dynamics simulations. The expectation of a broad distribution of helix orientations, at and above physiological ionic strength, is not met. Instead, this analysis shows that the HJH structures are dominated by two distinct conformations at moderate to high ionic strength. Atomic structures, selected from the molecular dynamics simulations, reveal strong base-base interactions in the junction that critically constrain the conformational space available to the HJH molecule and lead to a surprising re-extension at high salt. These results are corroborated by comparison with previous single-molecule fluorescence resonance energy transfer experiments on the same constructs.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

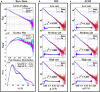
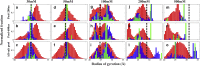
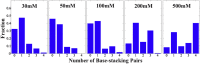
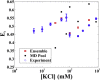

Similar articles
-
Refining the RNA Force Field with Small-Angle X-ray Scattering of Helix-Junction-Helix RNA.J Phys Chem Lett. 2022 Apr 21;13(15):3400-3408. doi: 10.1021/acs.jpclett.2c00359. Epub 2022 Apr 11. J Phys Chem Lett. 2022. PMID: 35404614 Free PMC article.
-
Tuning RNA Flexibility with Helix Length and Junction Sequence.Biophys J. 2015 Dec 15;109(12):2644-2653. doi: 10.1016/j.bpj.2015.10.039. Biophys J. 2015. PMID: 26682821 Free PMC article.
-
Visualizing RNA Structures by SAXS-Driven MD Simulations.Front Bioinform. 2022 Feb 18;2:781949. doi: 10.3389/fbinf.2022.781949. eCollection 2022. Front Bioinform. 2022. PMID: 36304317 Free PMC article.
-
Hybrid Methods for Modeling Protein Structures Using Molecular Dynamics Simulations and Small-Angle X-Ray Scattering Data.Adv Exp Med Biol. 2018;1105:237-258. doi: 10.1007/978-981-13-2200-6_15. Adv Exp Med Biol. 2018. PMID: 30617833 Review.
-
SAXS studies of RNA: structures, dynamics, and interactions with partners.Wiley Interdiscip Rev RNA. 2016 Jul;7(4):512-26. doi: 10.1002/wrna.1349. Epub 2016 Apr 12. Wiley Interdiscip Rev RNA. 2016. PMID: 27071649 Free PMC article. Review.
Cited by
-
Refining the RNA Force Field with Small-Angle X-ray Scattering of Helix-Junction-Helix RNA.J Phys Chem Lett. 2022 Apr 21;13(15):3400-3408. doi: 10.1021/acs.jpclett.2c00359. Epub 2022 Apr 11. J Phys Chem Lett. 2022. PMID: 35404614 Free PMC article.
-
Structure folding of RNA kissing complexes in salt solutions: predicting 3D structure, stability, and folding pathway.RNA. 2019 Nov;25(11):1532-1548. doi: 10.1261/rna.071662.119. Epub 2019 Aug 7. RNA. 2019. PMID: 31391217 Free PMC article.
-
Assembly and Stability of Simian Virus 40 Polymorphs.ACS Nano. 2020 Apr 28;14(4):4430-4443. doi: 10.1021/acsnano.9b10004. Epub 2020 Apr 2. ACS Nano. 2020. PMID: 32208635 Free PMC article.
-
Predicting RNA Structure and Dynamics with Deep Learning and Solution Scattering.bioRxiv [Preprint]. 2024 Dec 21:2024.06.08.598075. doi: 10.1101/2024.06.08.598075. bioRxiv. 2024. Update in: Biophys J. 2025 Feb 04;124(3):549-564. doi: 10.1016/j.bpj.2024.12.024. PMID: 39764023 Free PMC article. Updated. Preprint.
-
Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.J Am Chem Soc. 2023 Oct 25;145(42):22964-22978. doi: 10.1021/jacs.3c04614. Epub 2023 Oct 13. J Am Chem Soc. 2023. PMID: 37831584 Free PMC article.
References
-
- Amara S.G., Jonas V., Evans R.M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. - PubMed
-
- McCaffrey A.P., Meuse L., Kay M.A. RNA interference in adult mice. Nature. 2002;418:38–39. - PubMed
-
- Jackson A.L., Bartz S.R., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. - PubMed
-
- Tucker B.J., Breaker R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005;15:342–348. - PubMed
-
- Robertson D.L., Joyce G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

